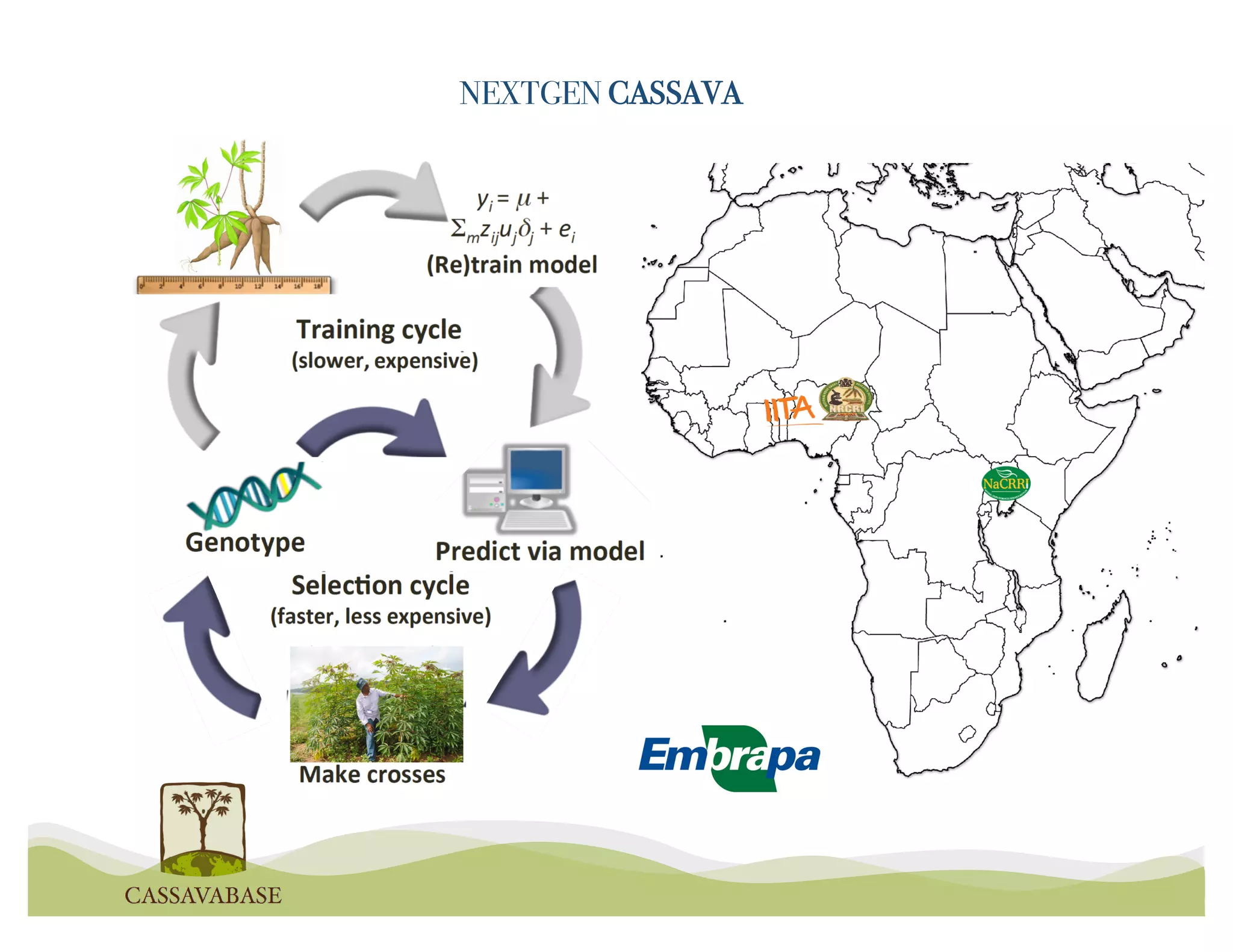
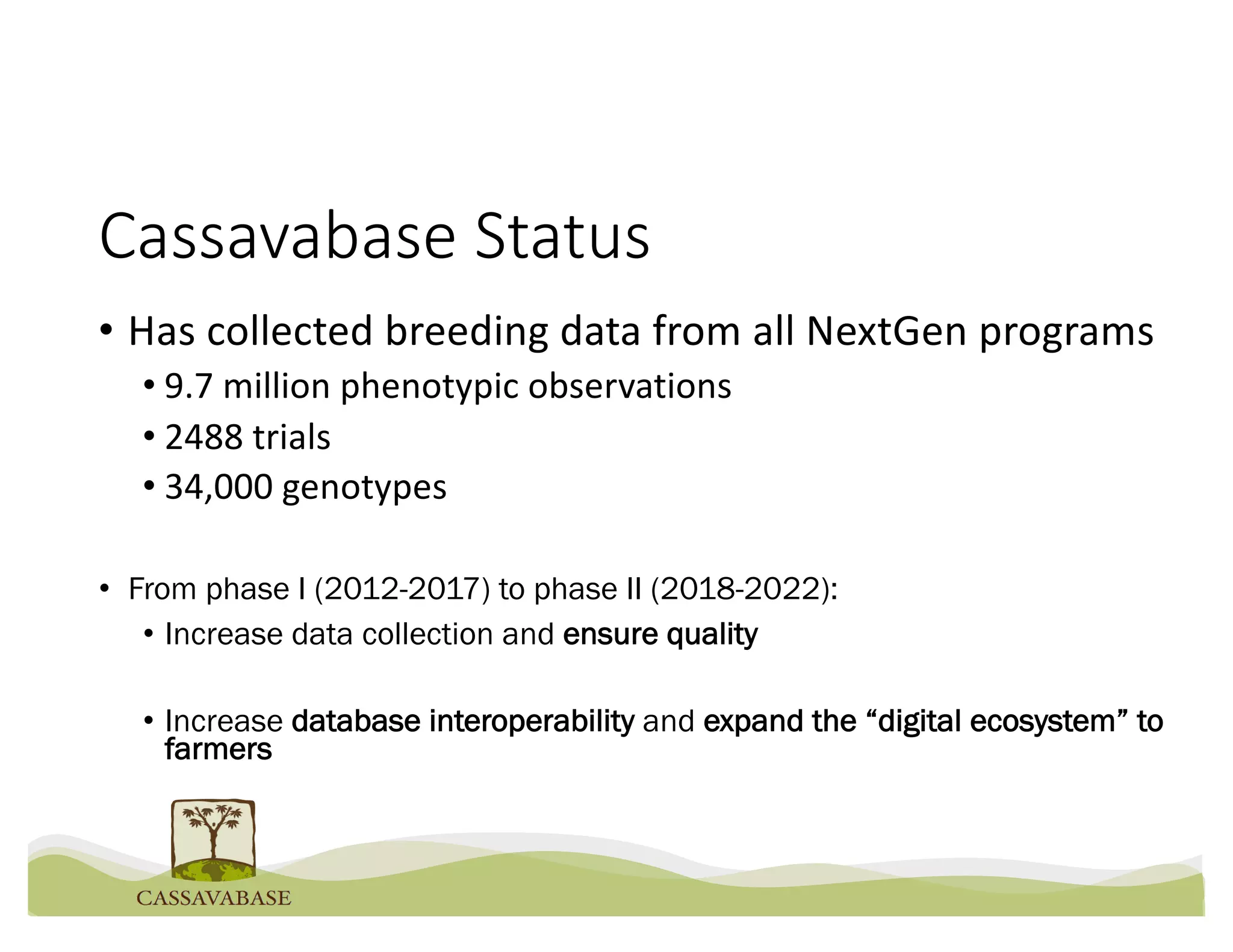
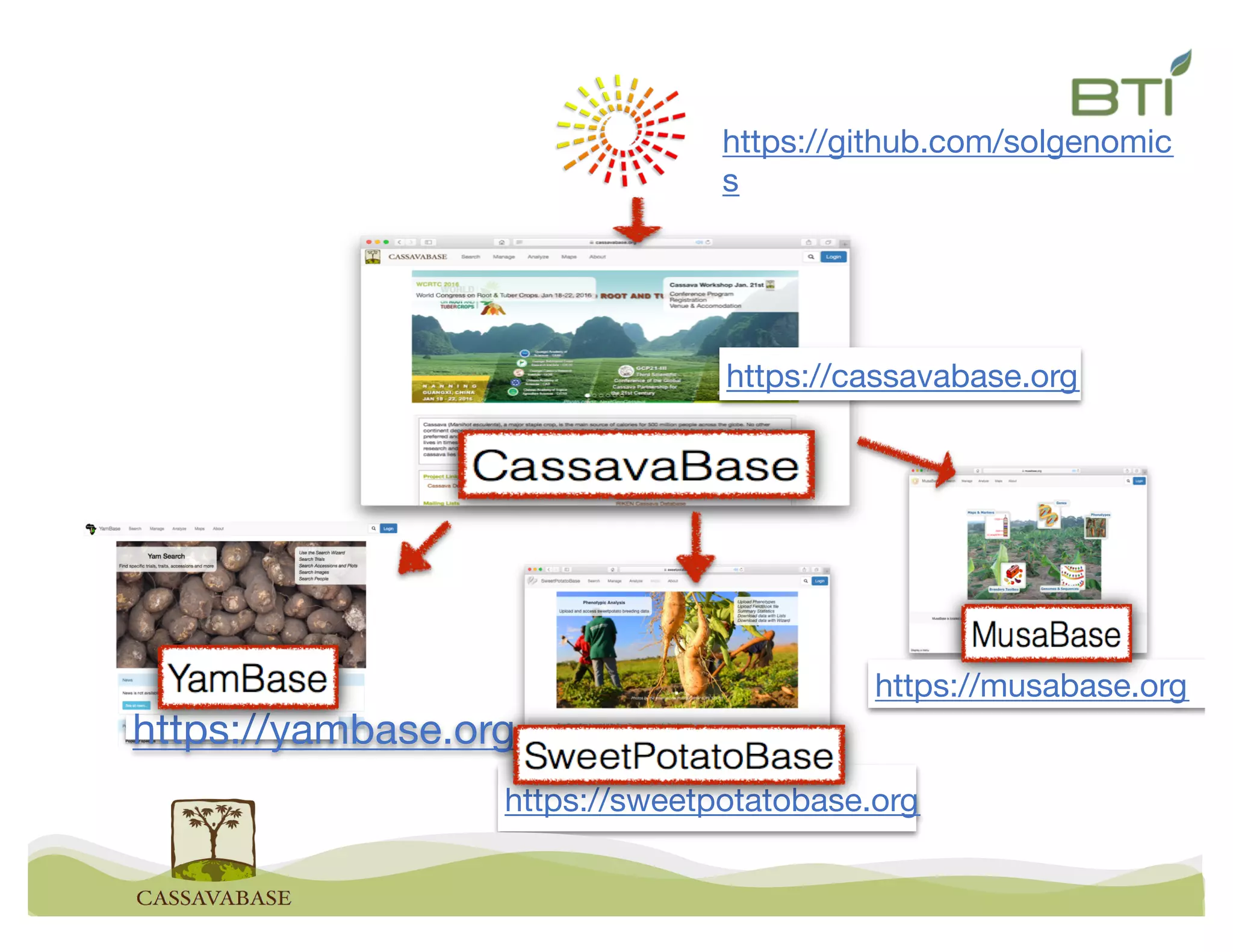
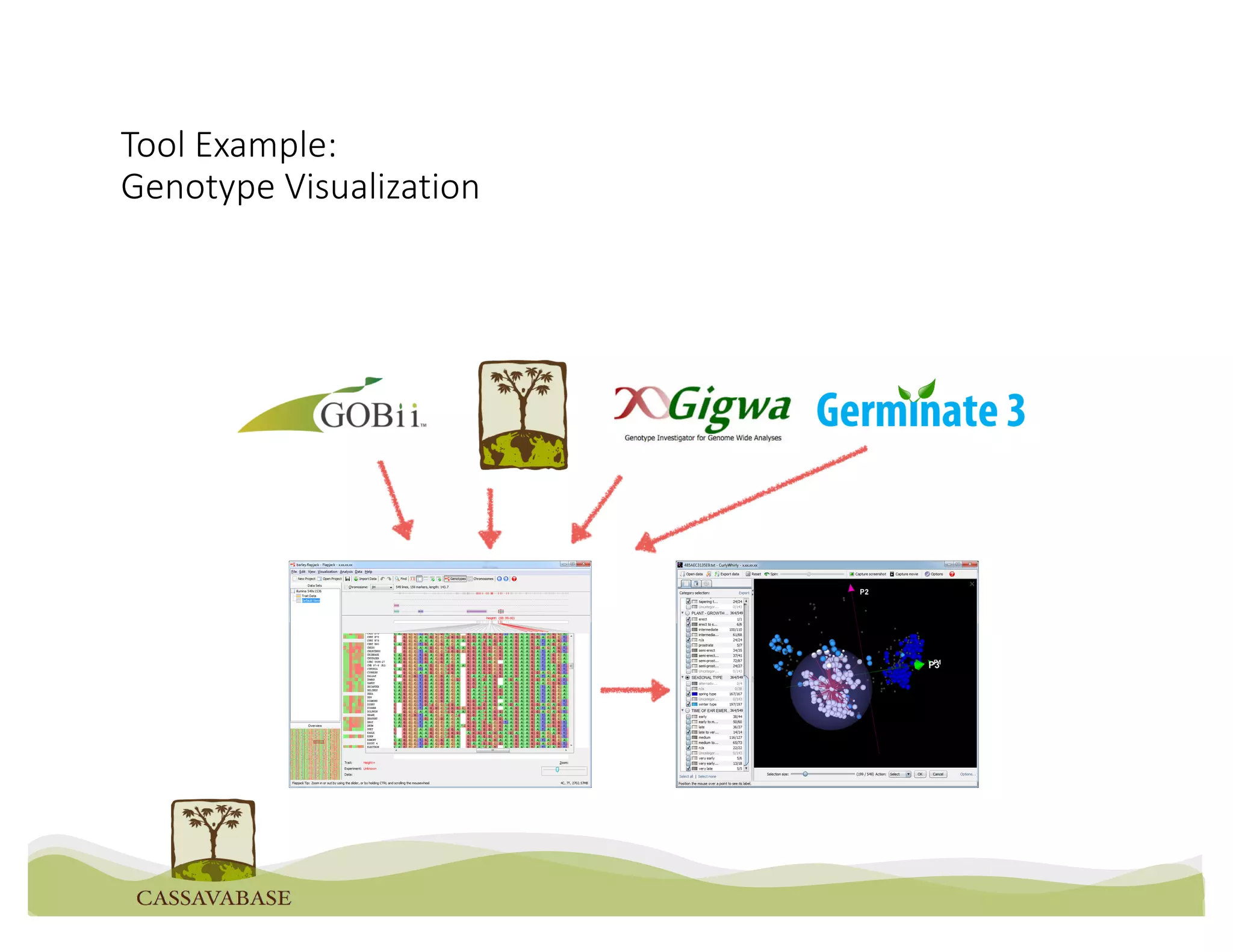
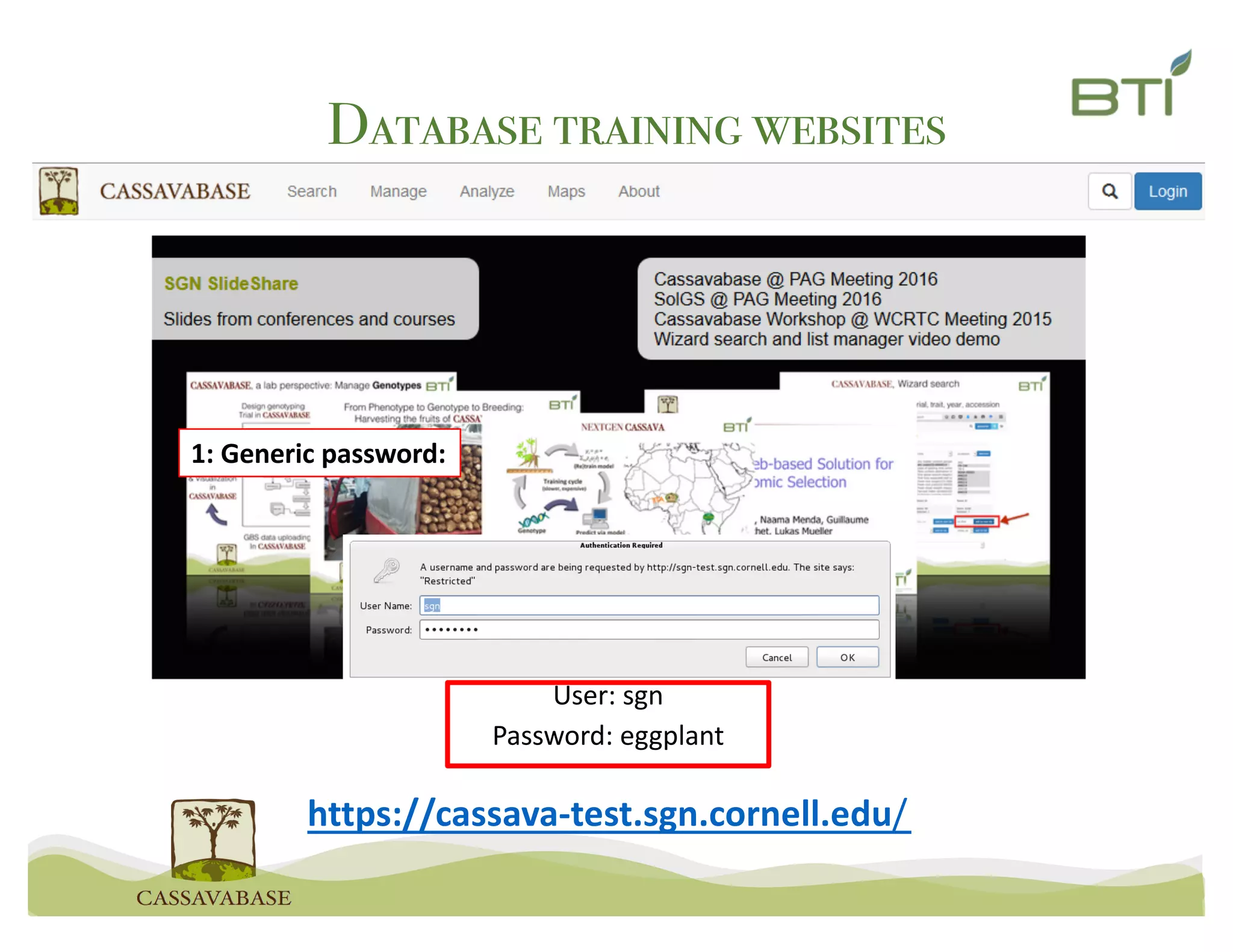
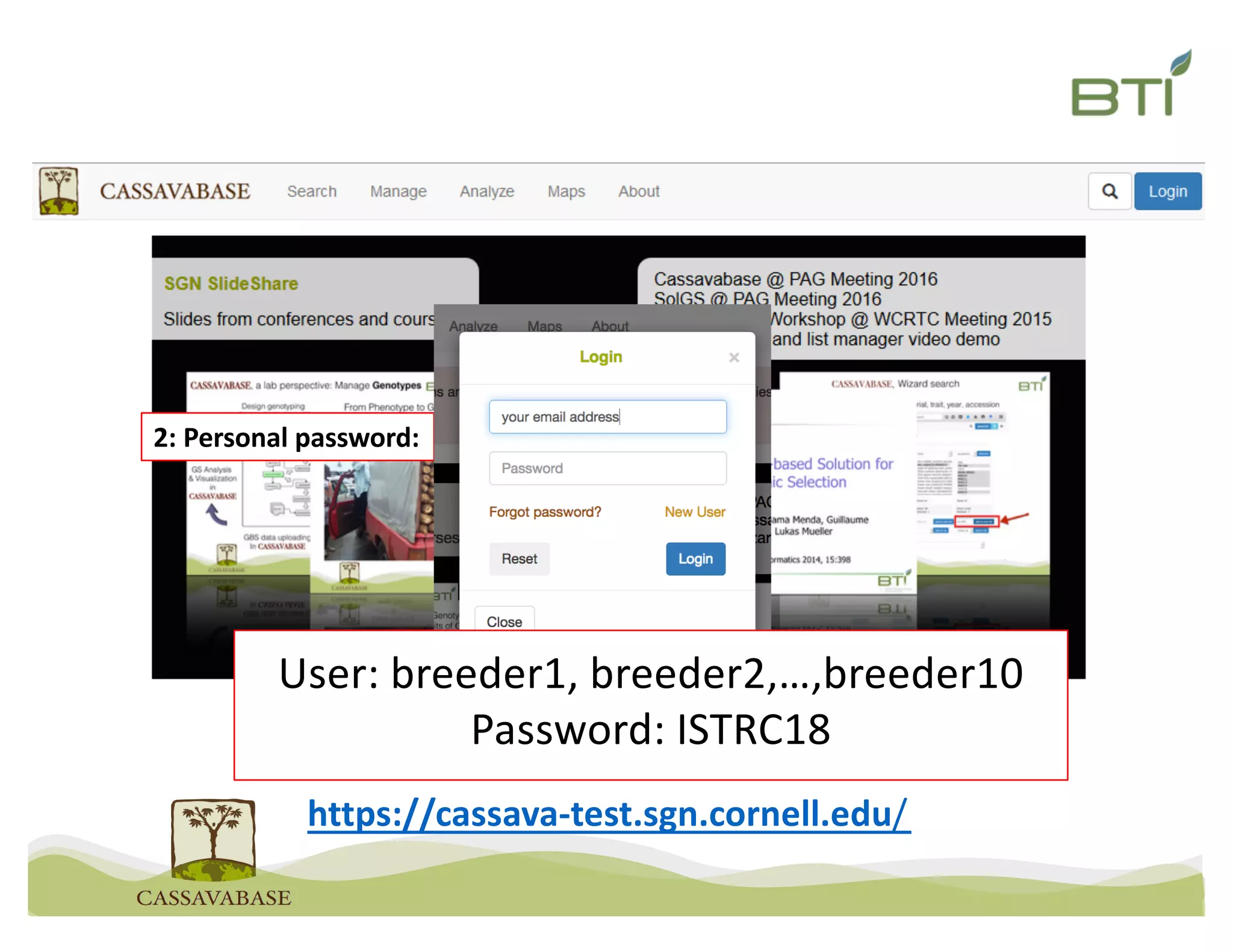
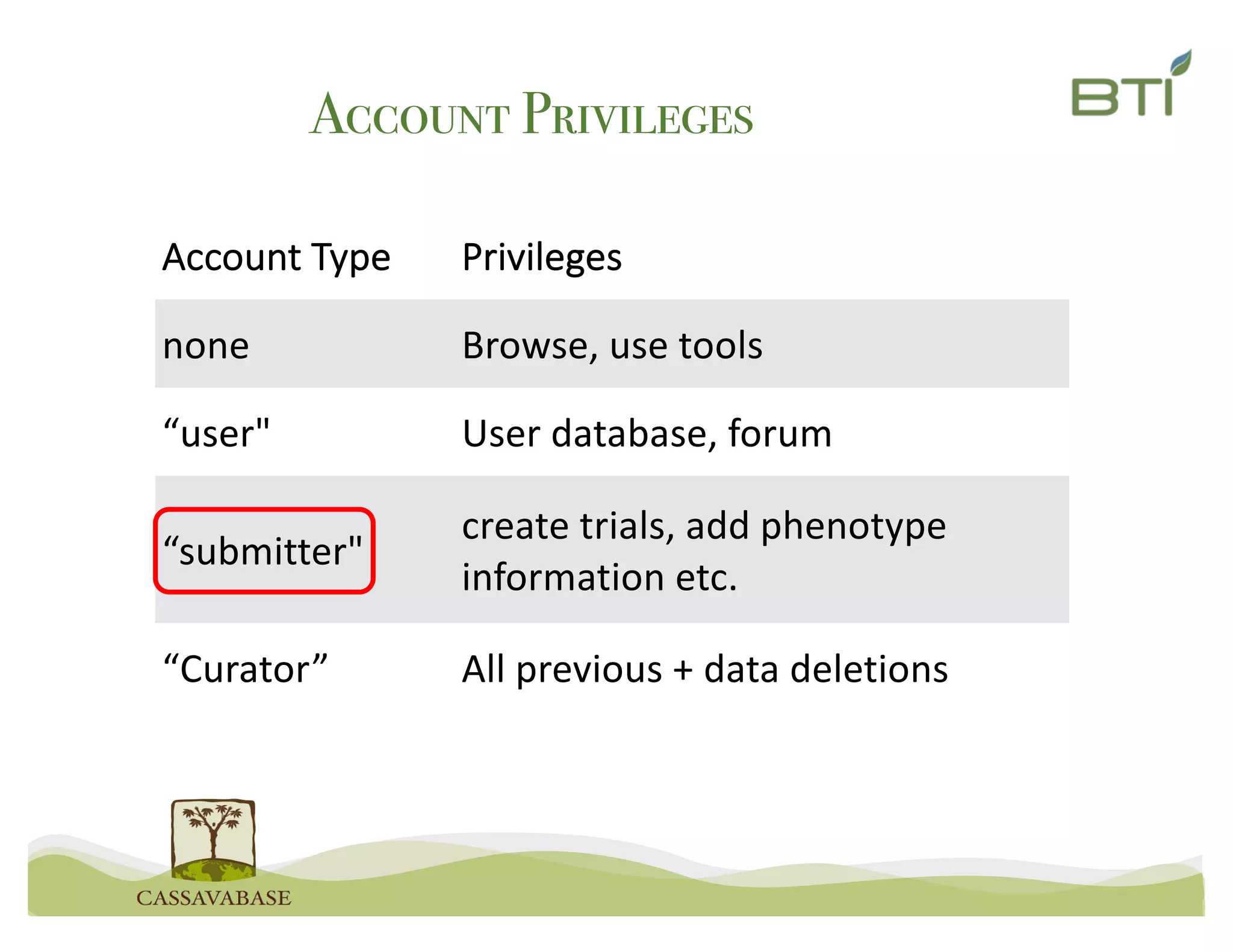
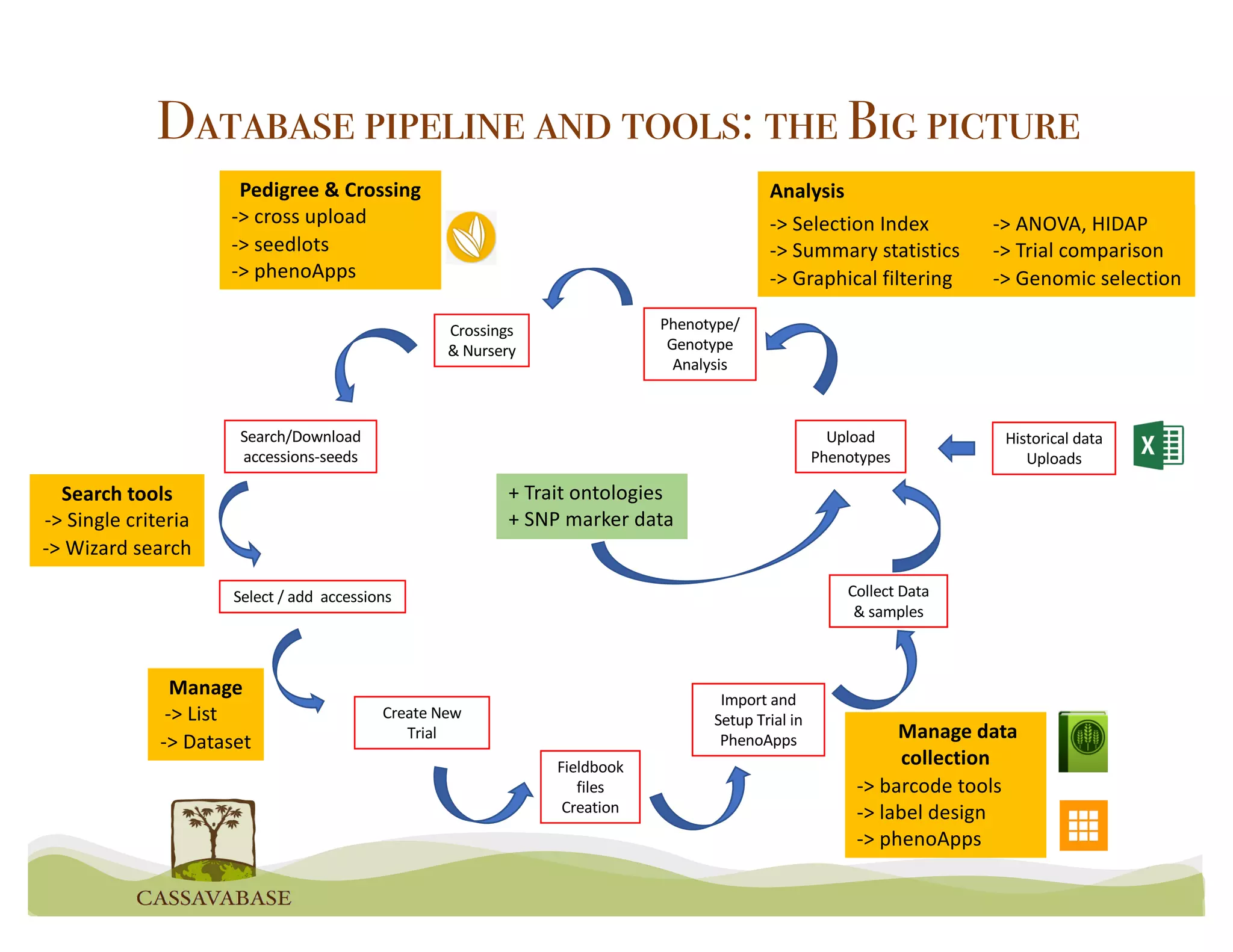
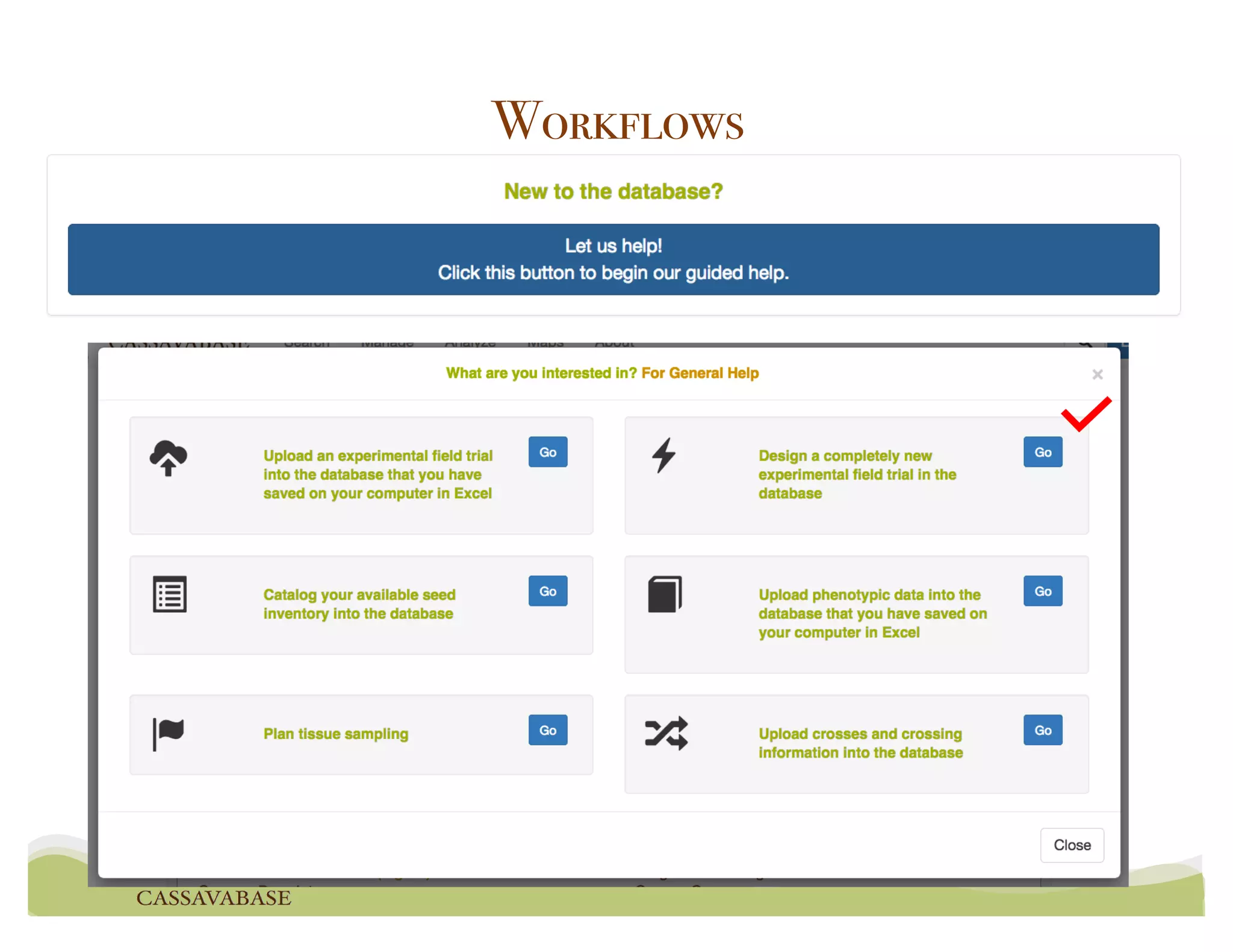
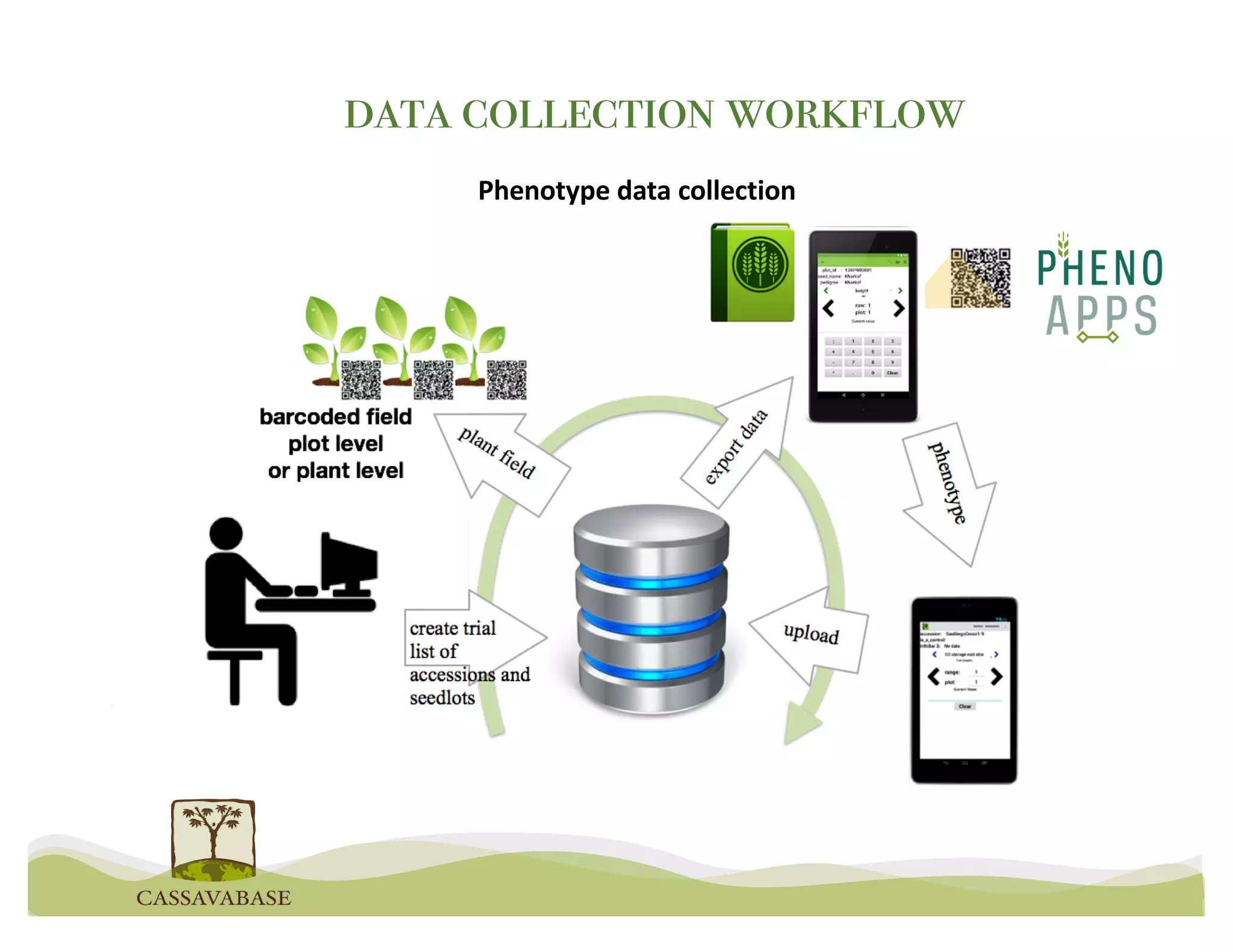
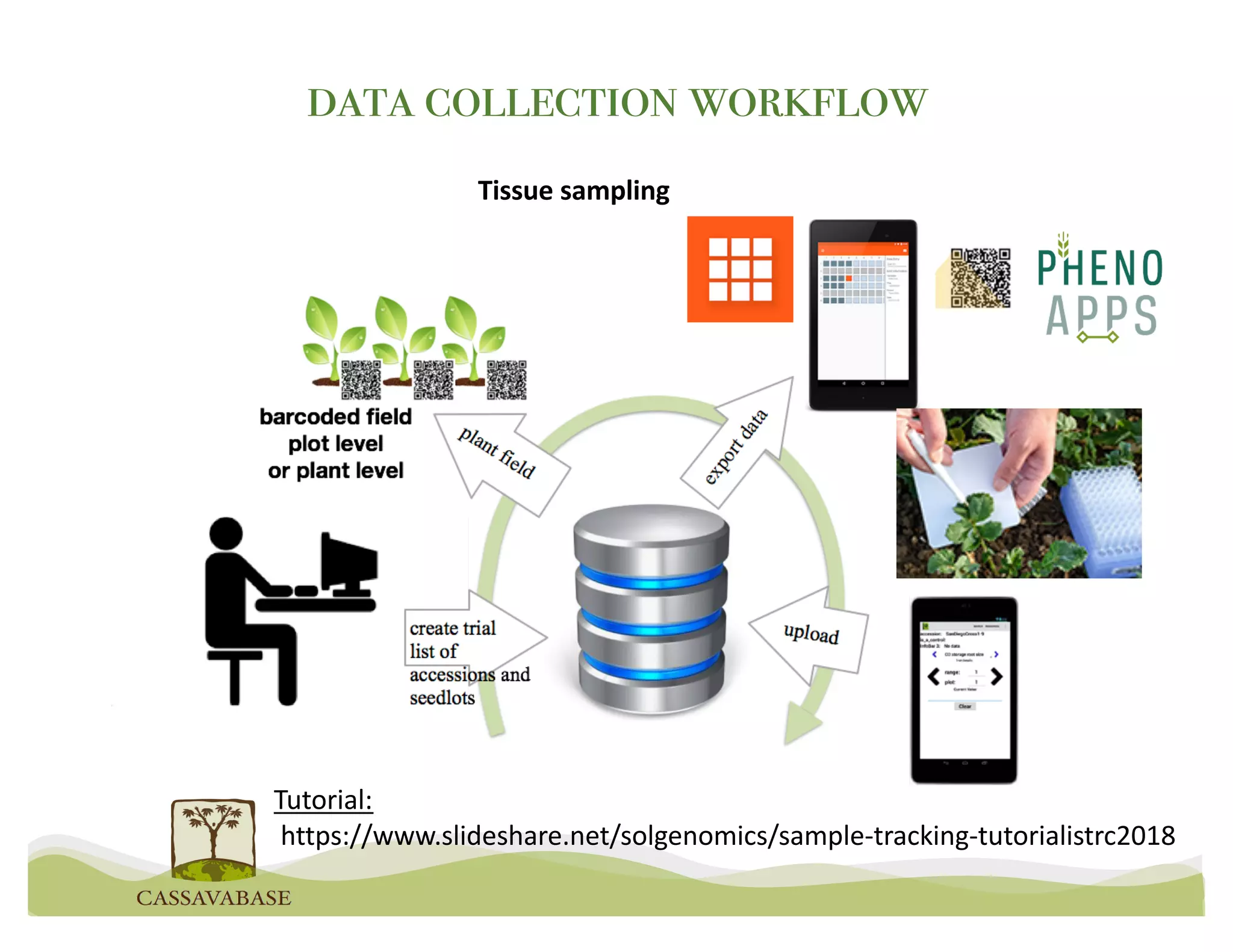
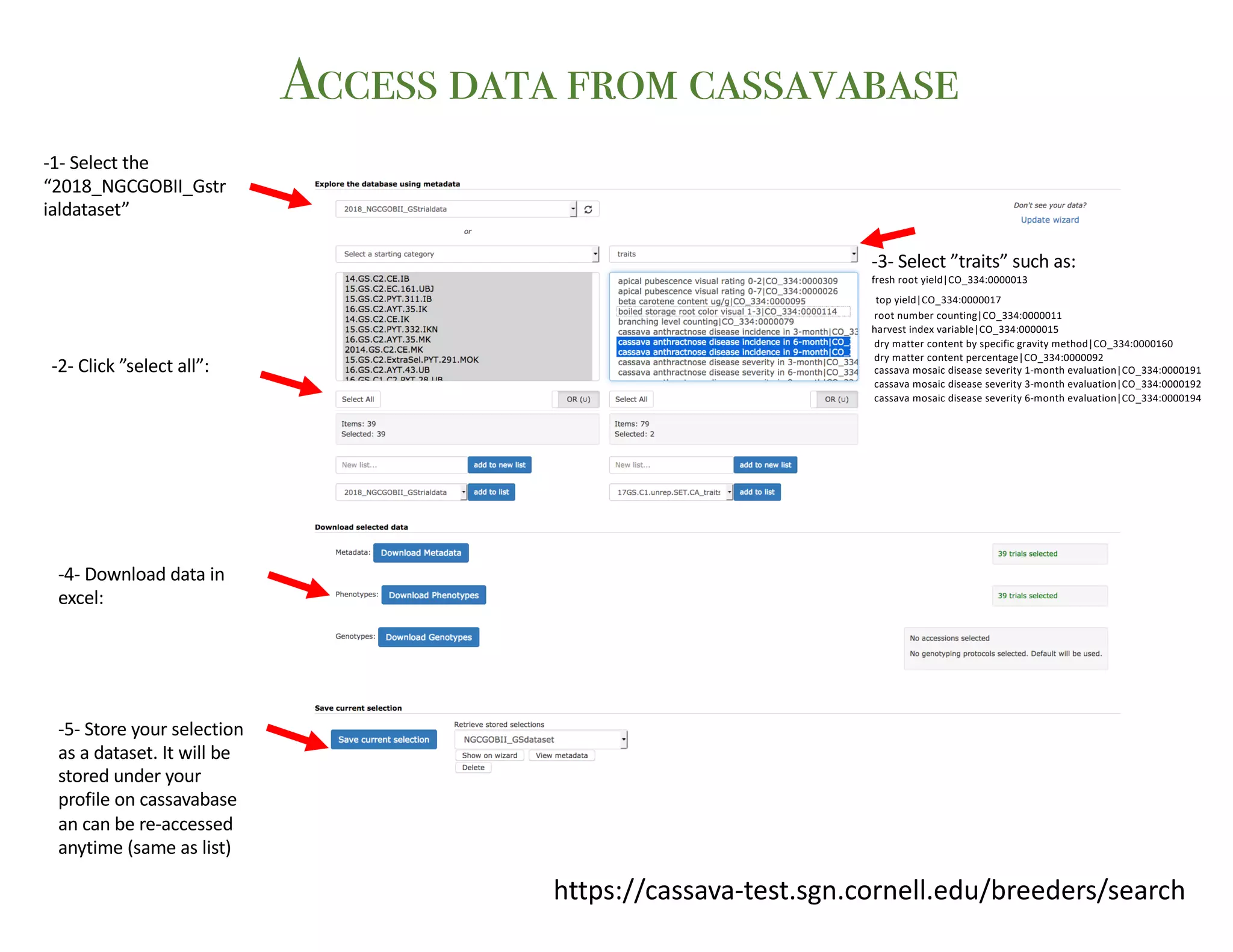
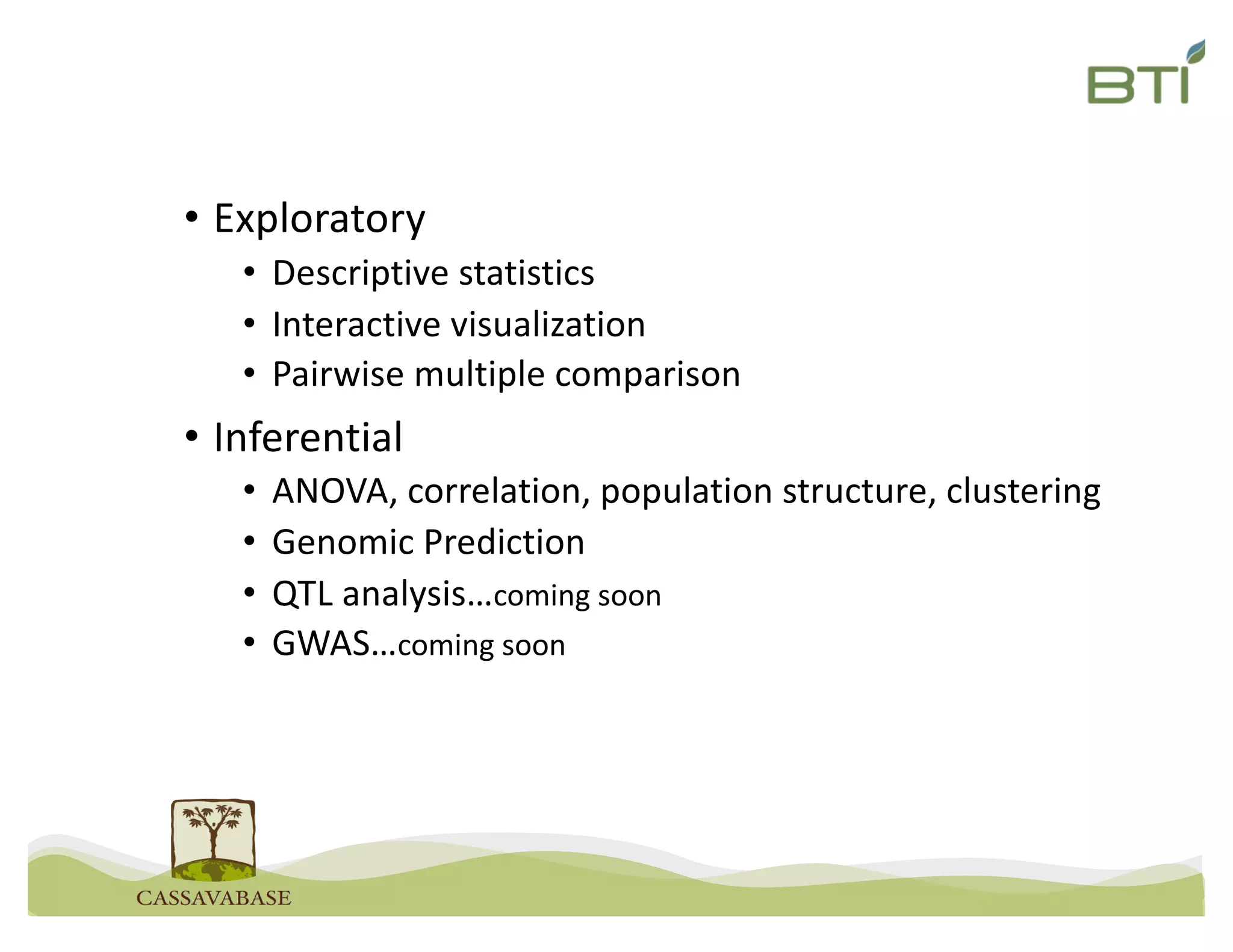
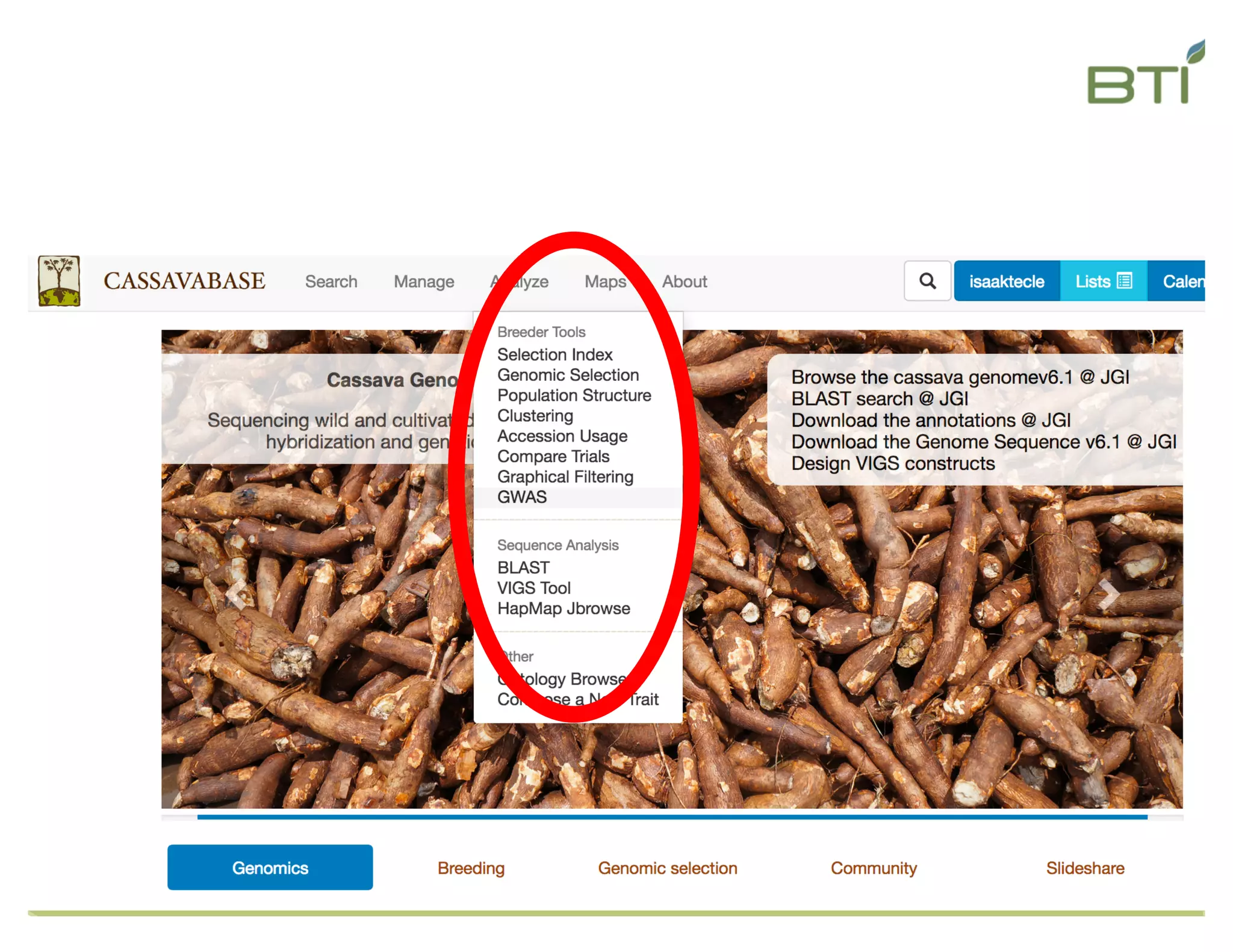
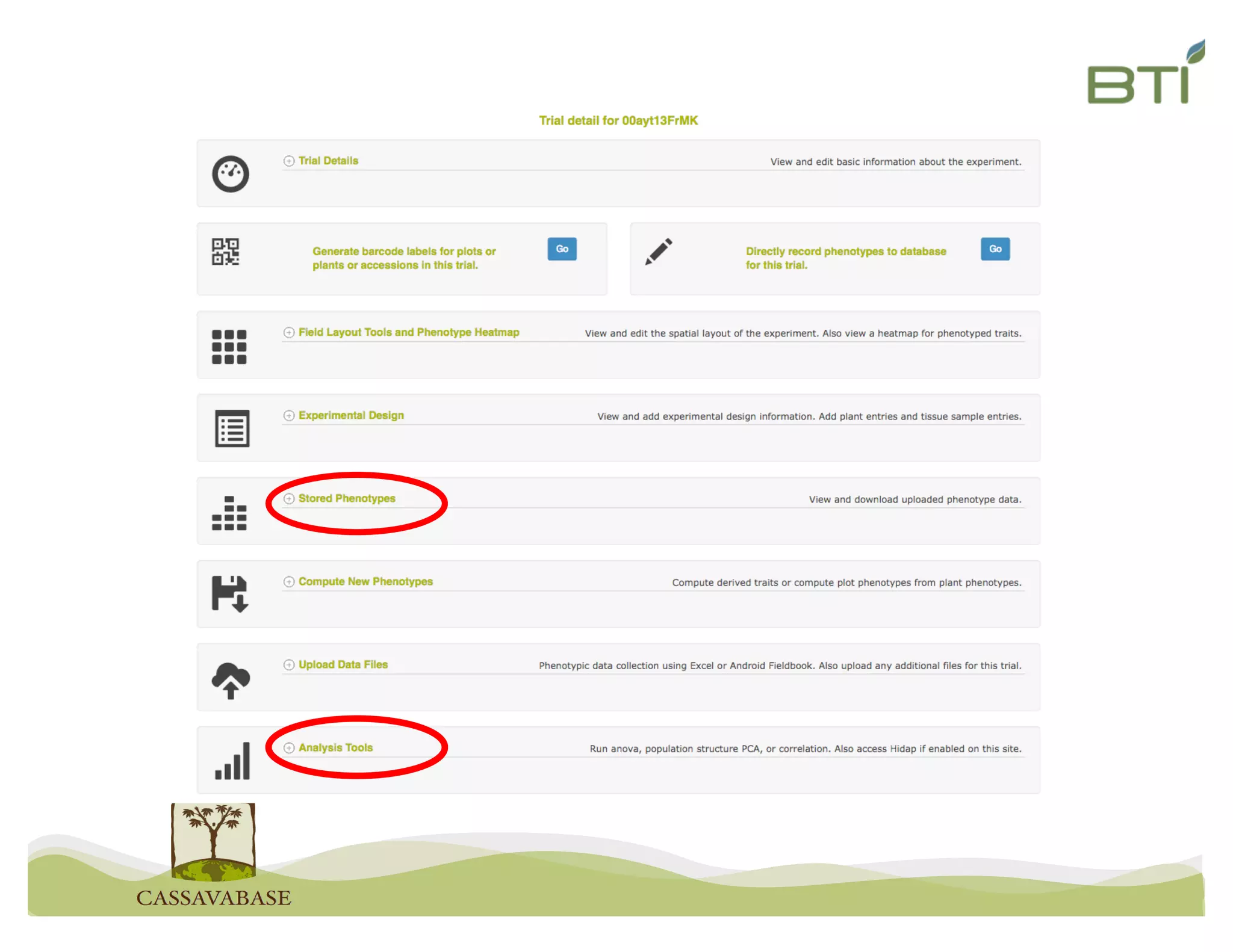
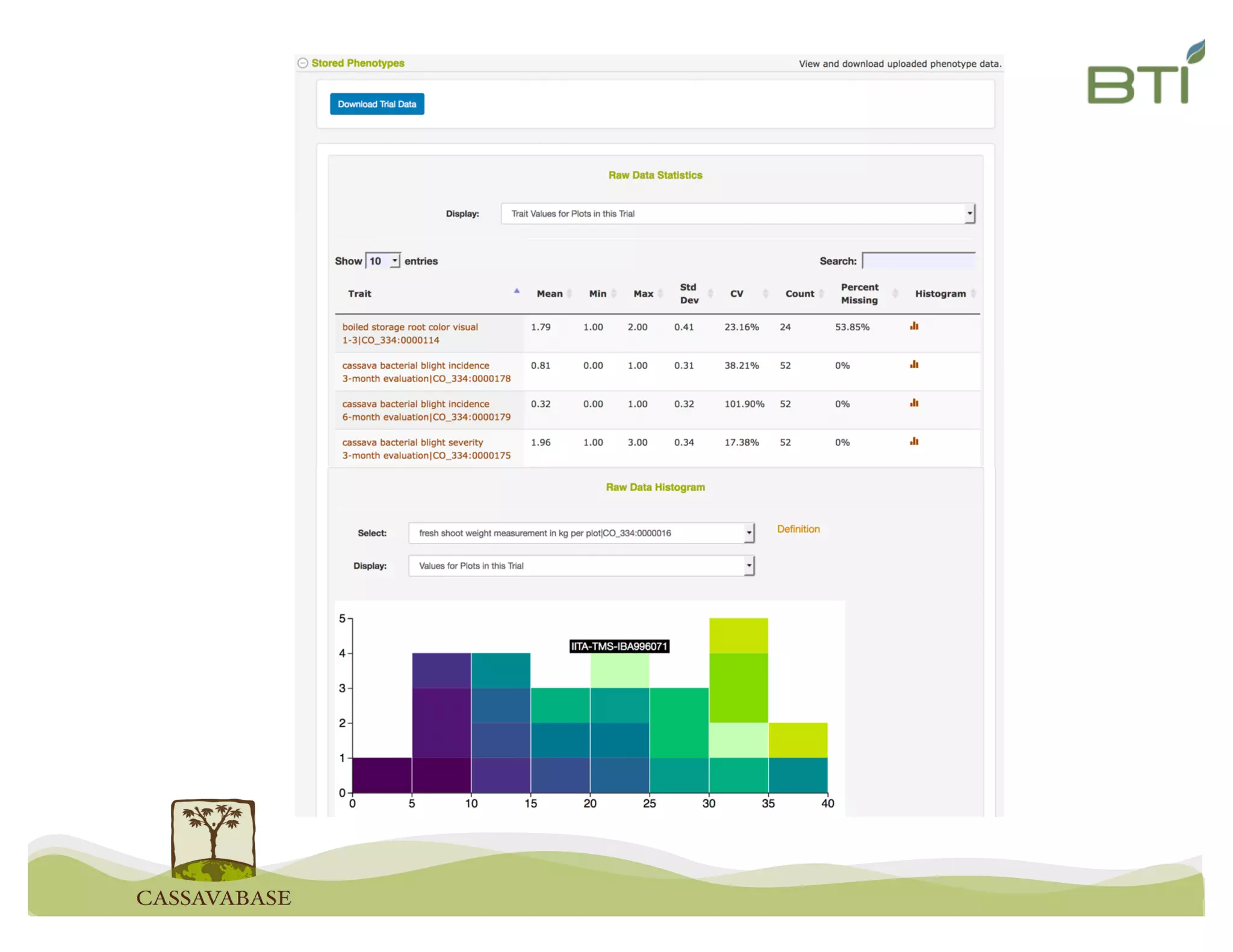
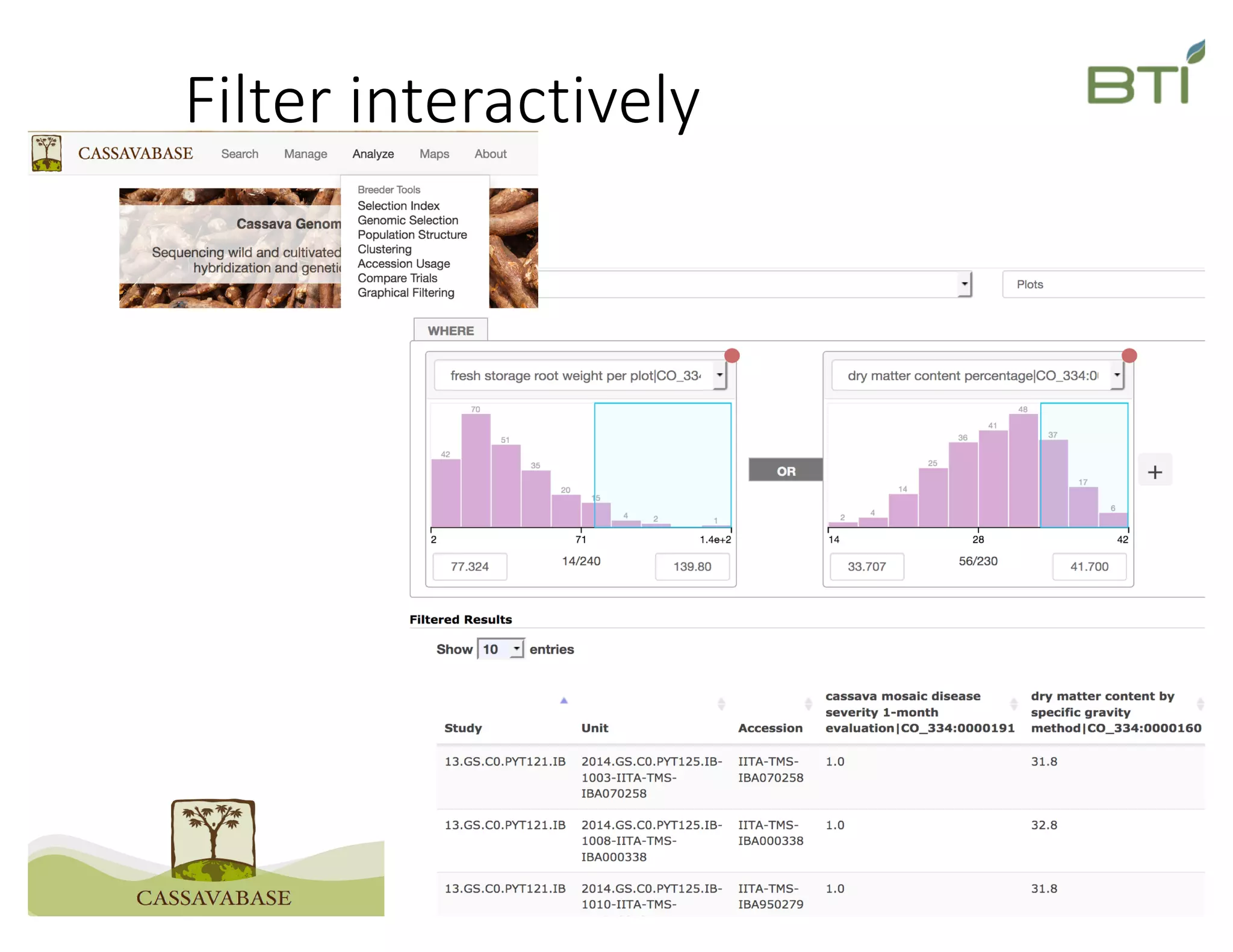
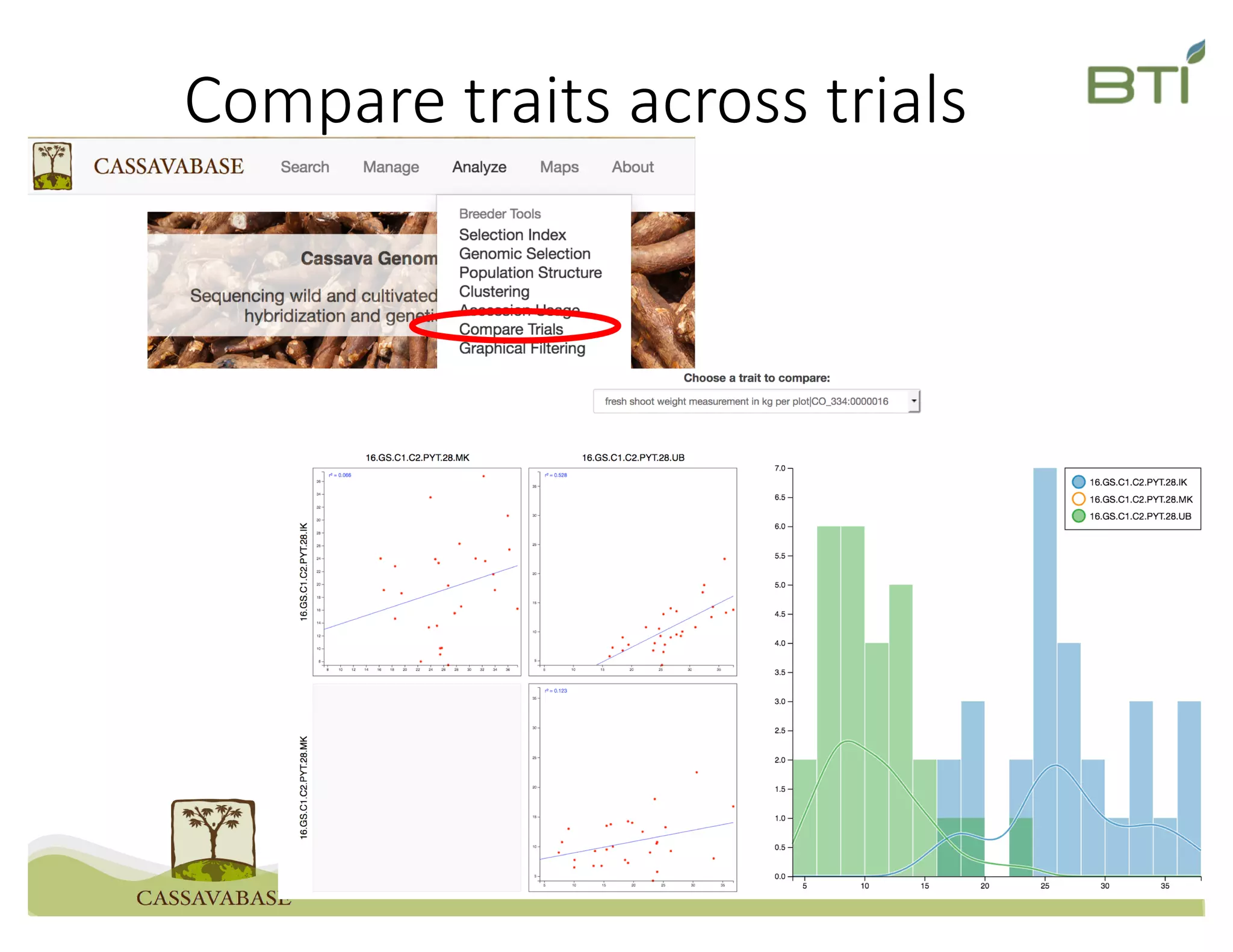
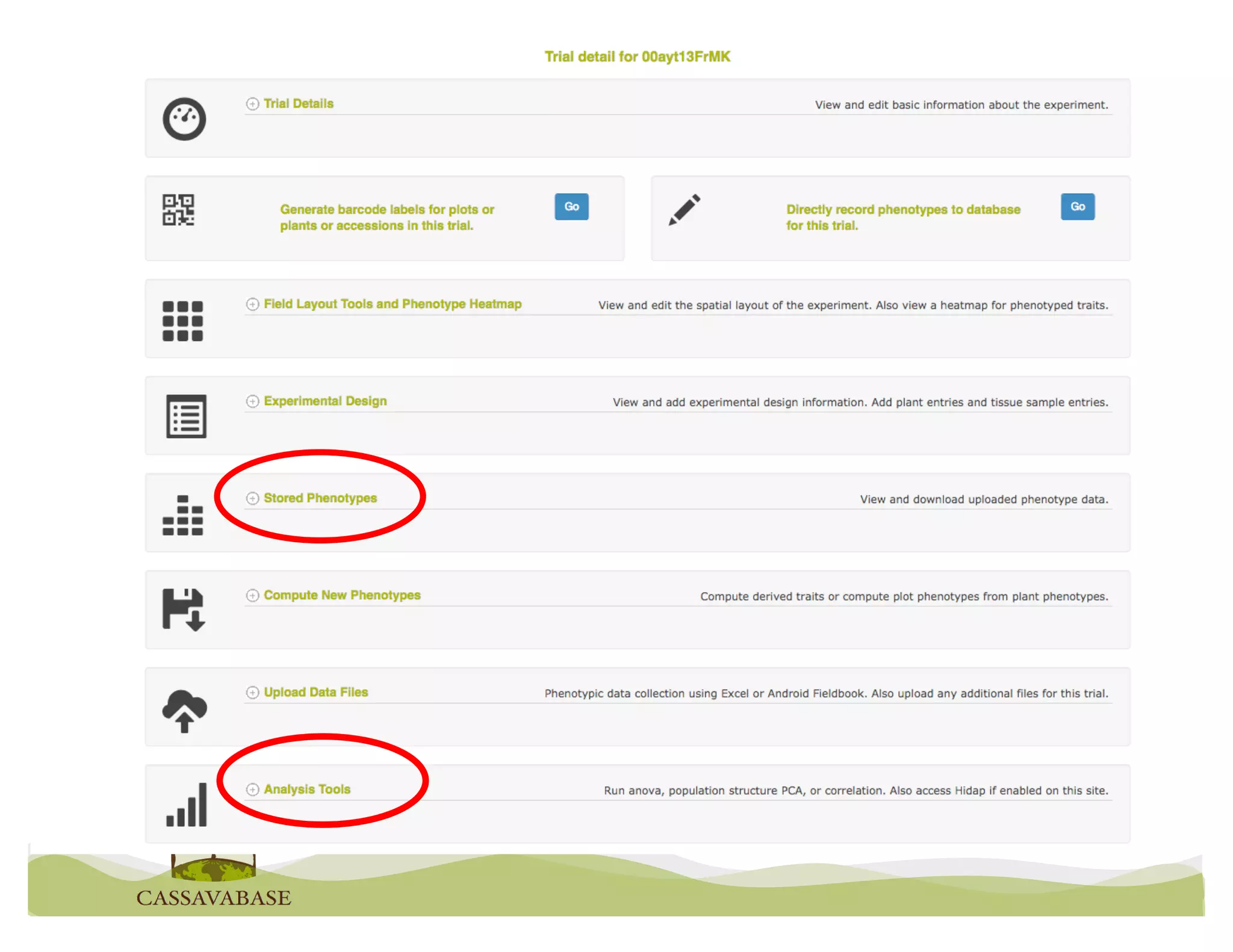
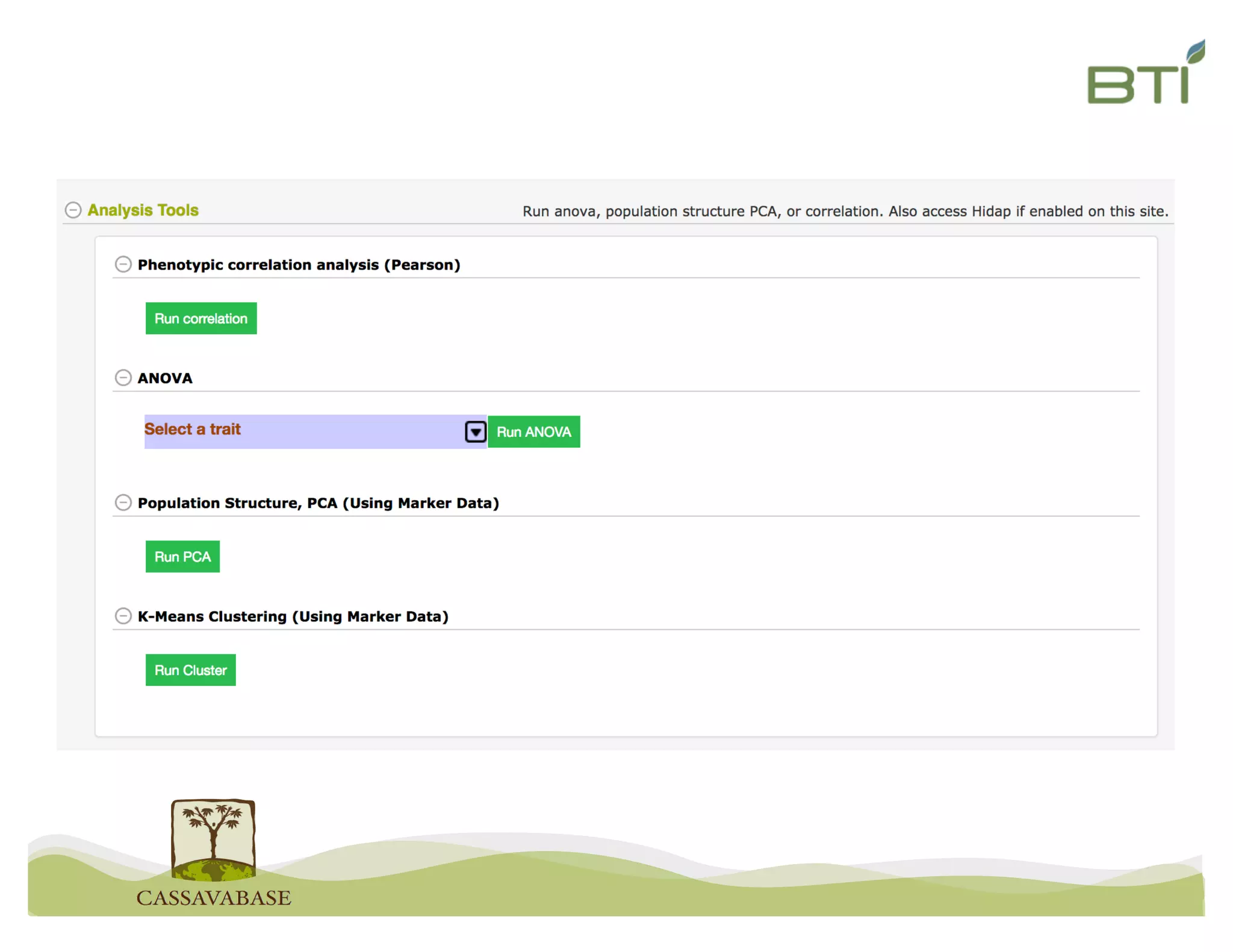
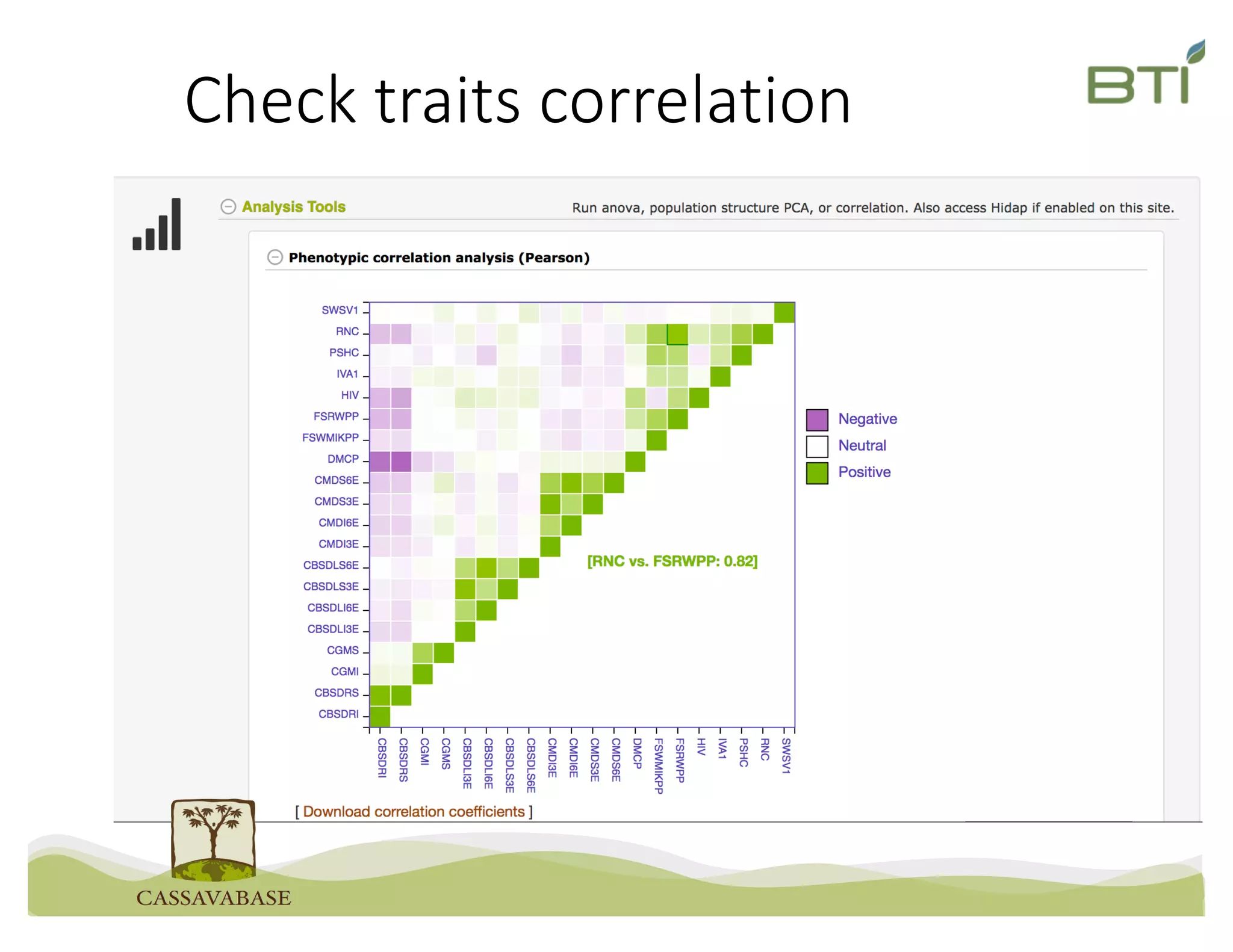
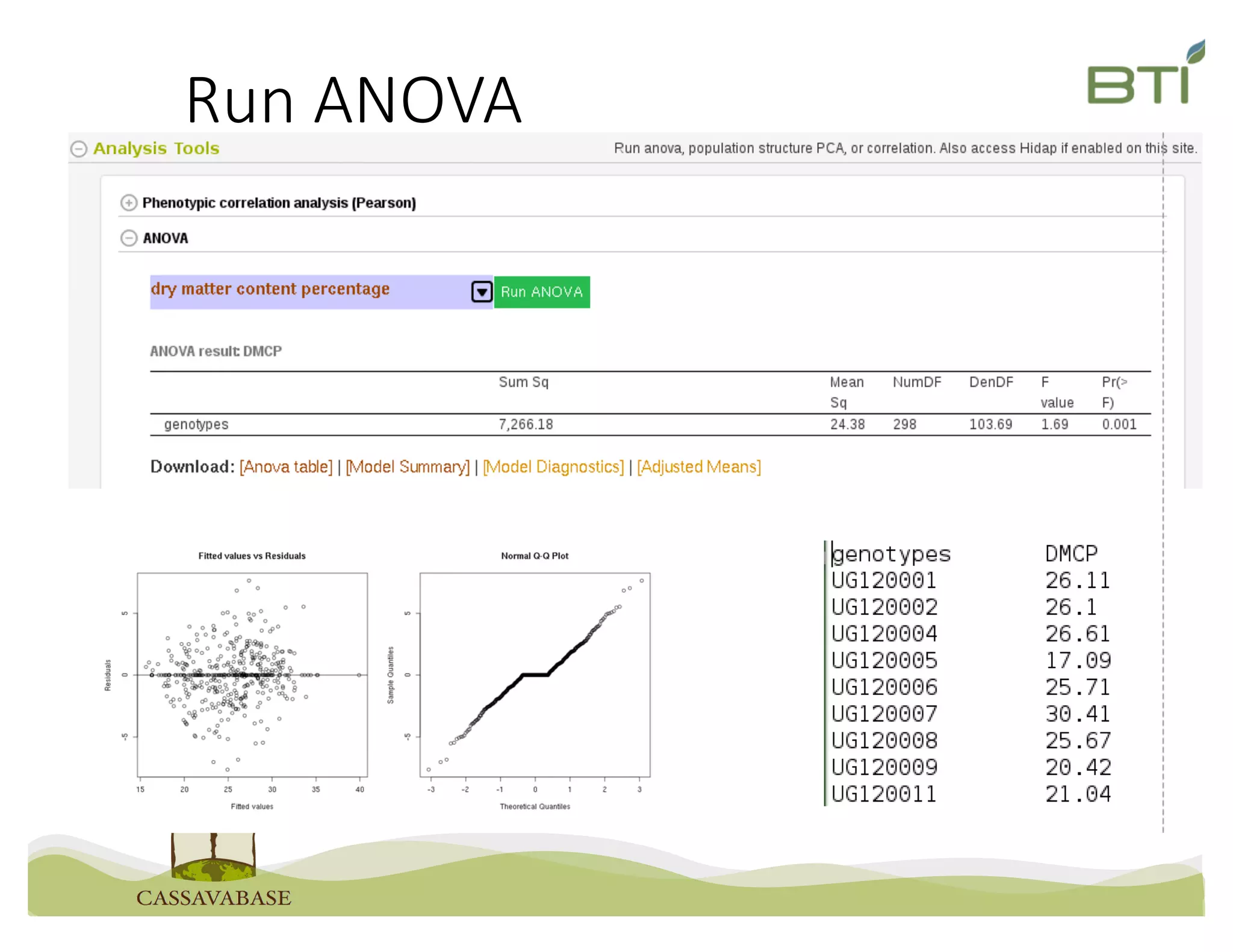
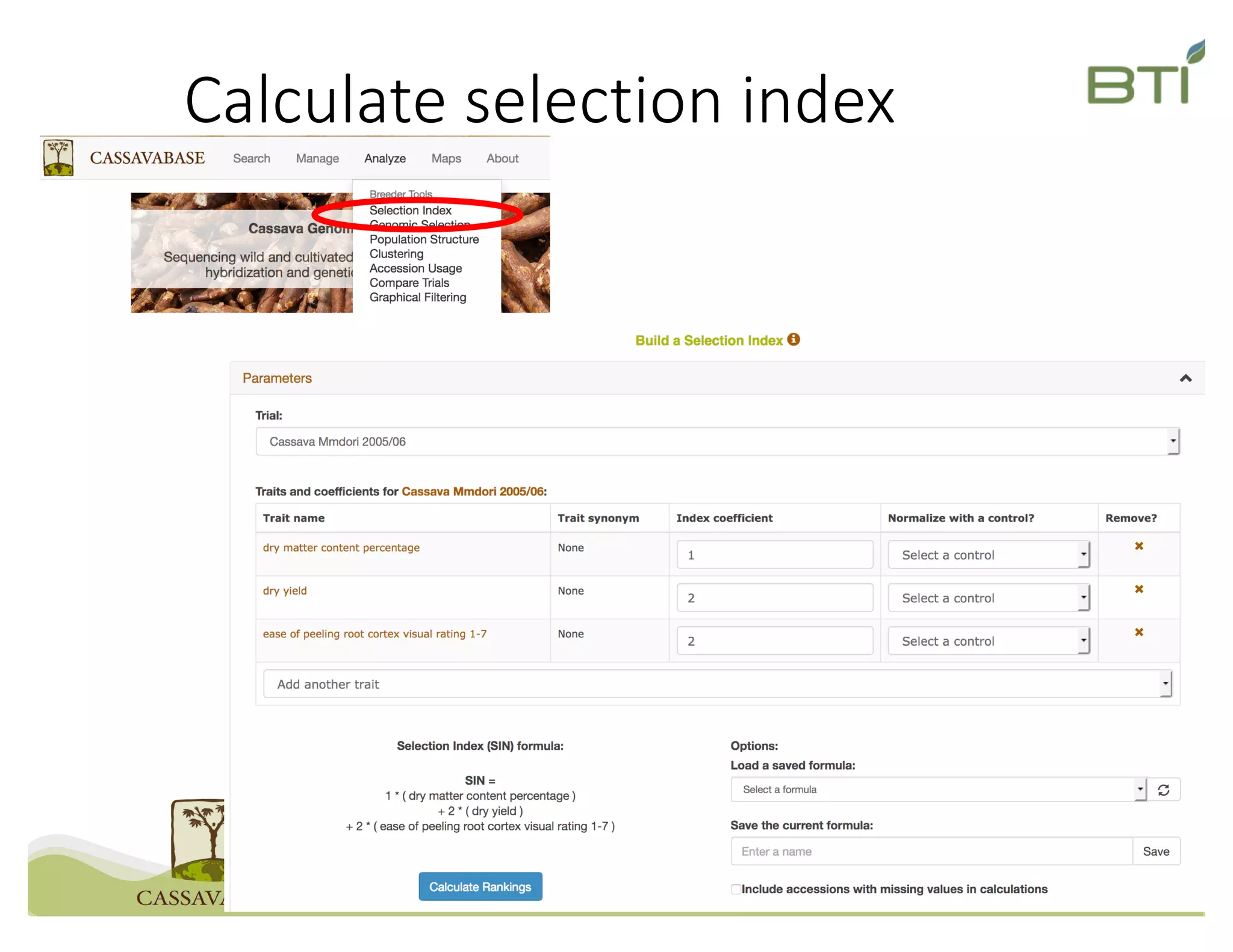
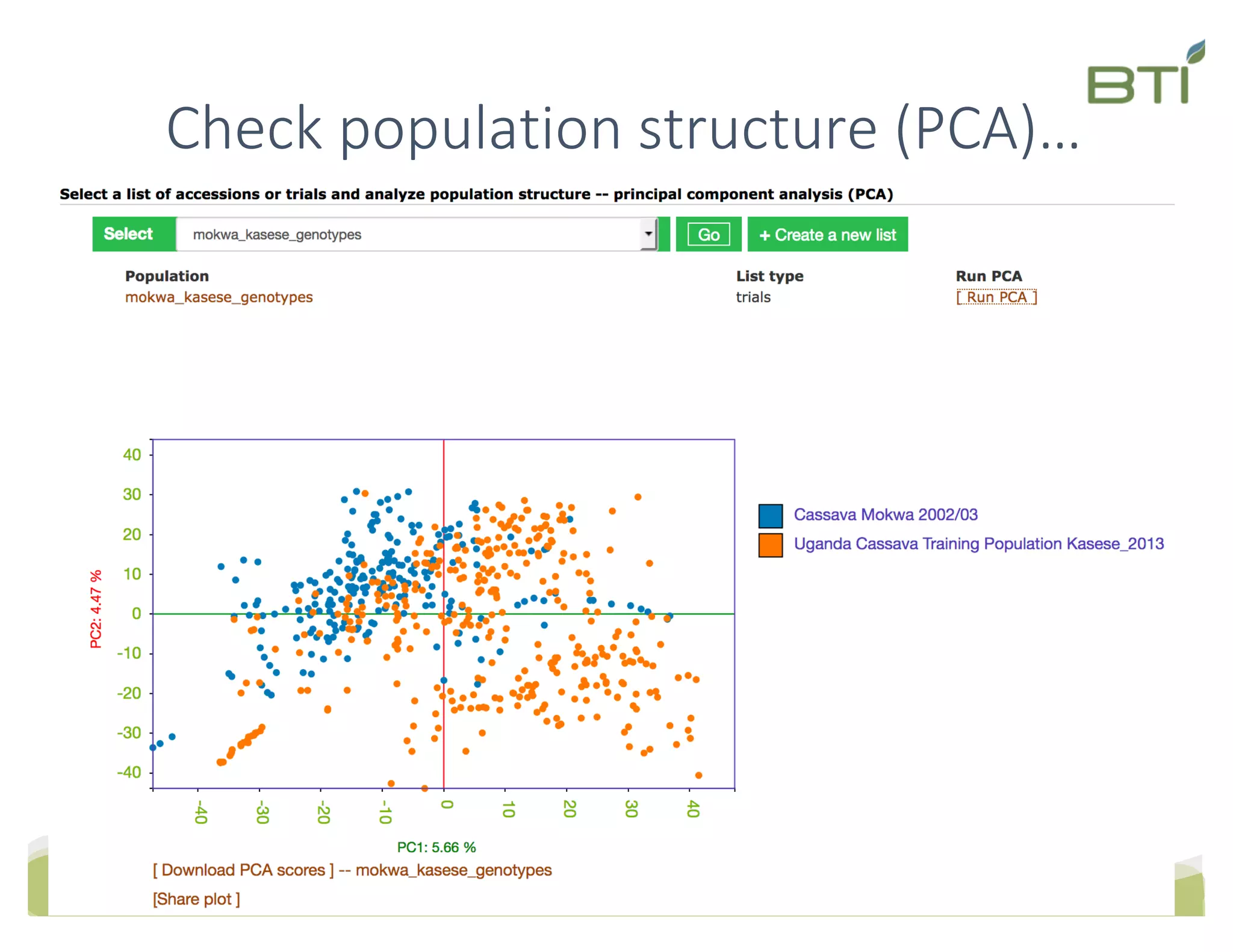
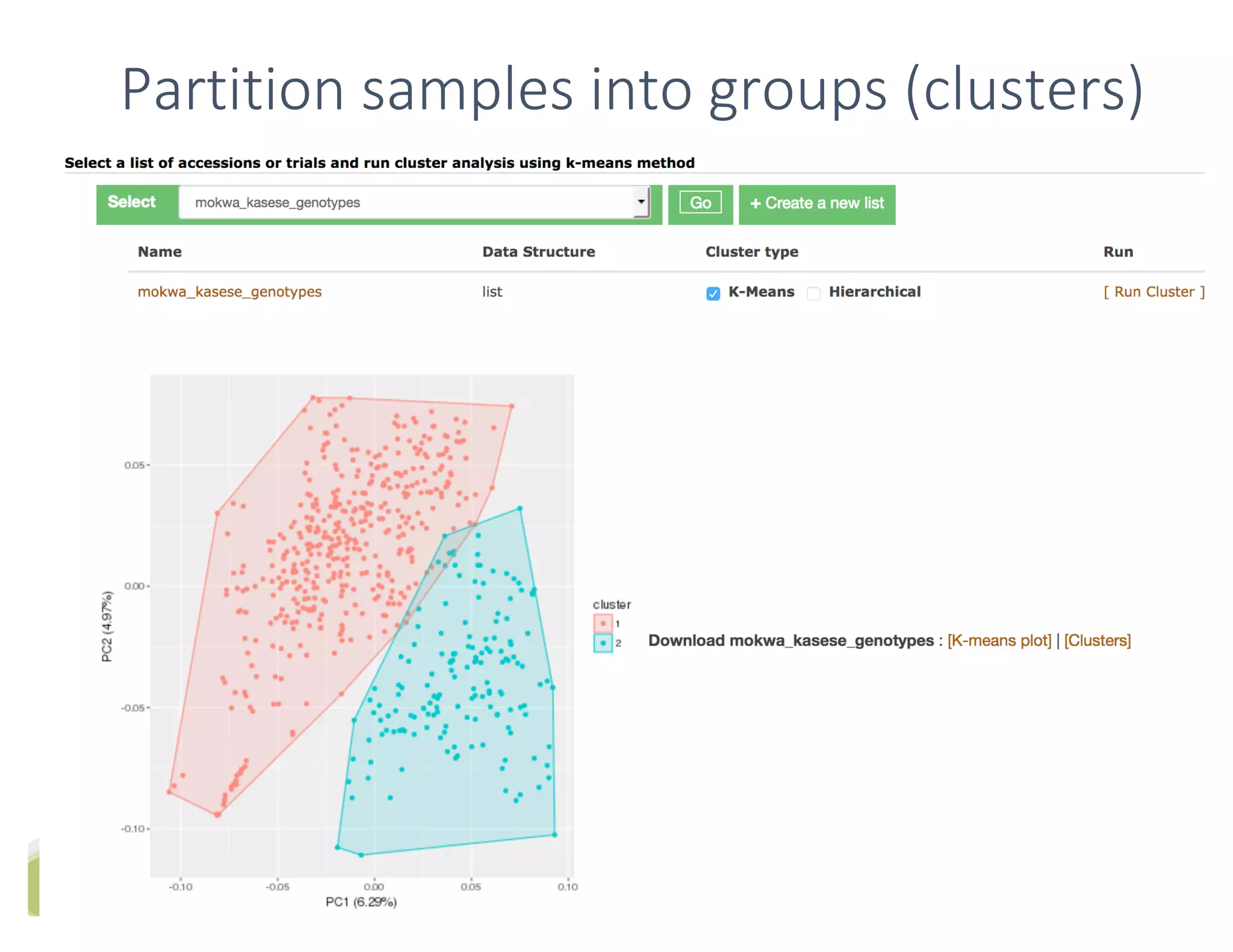
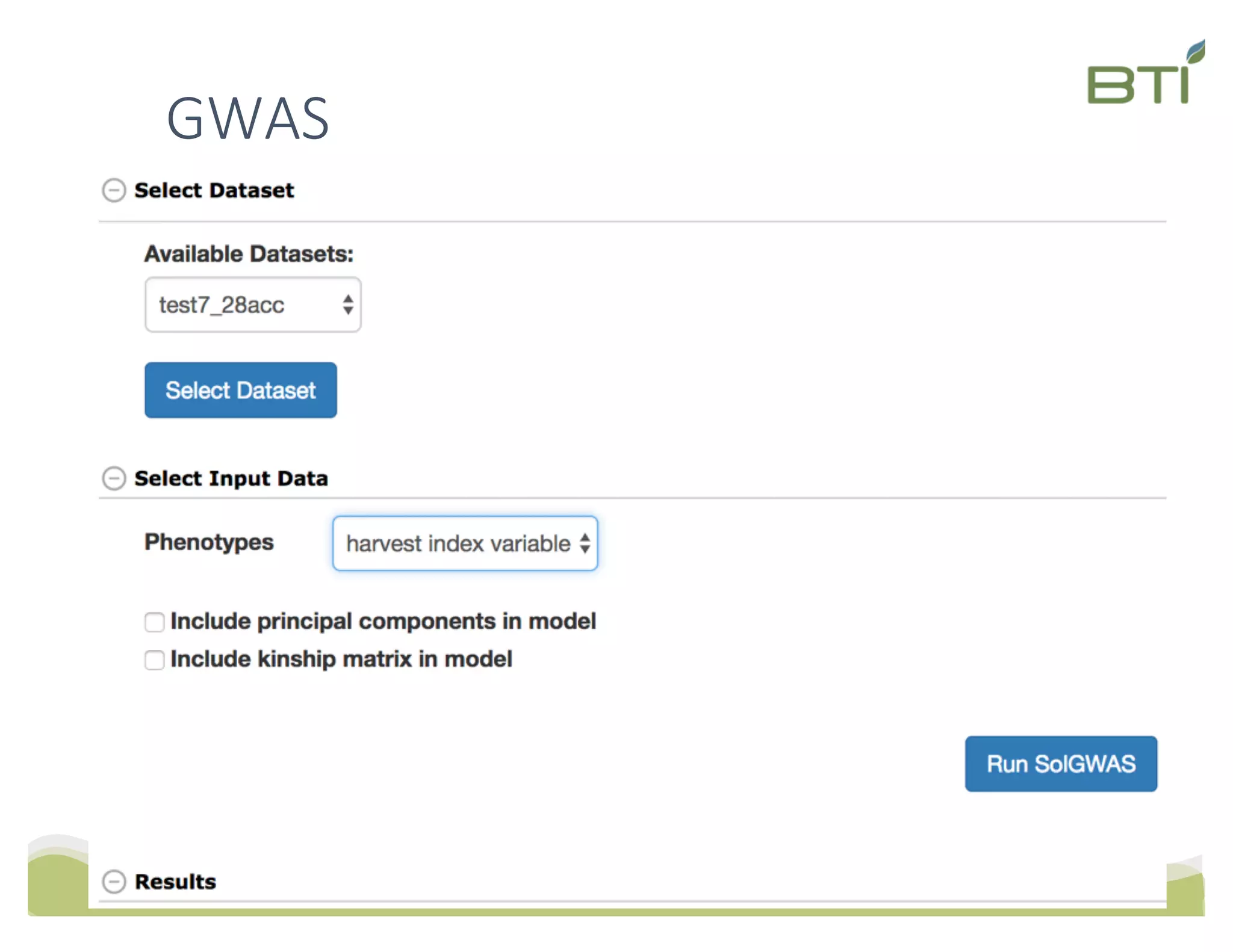
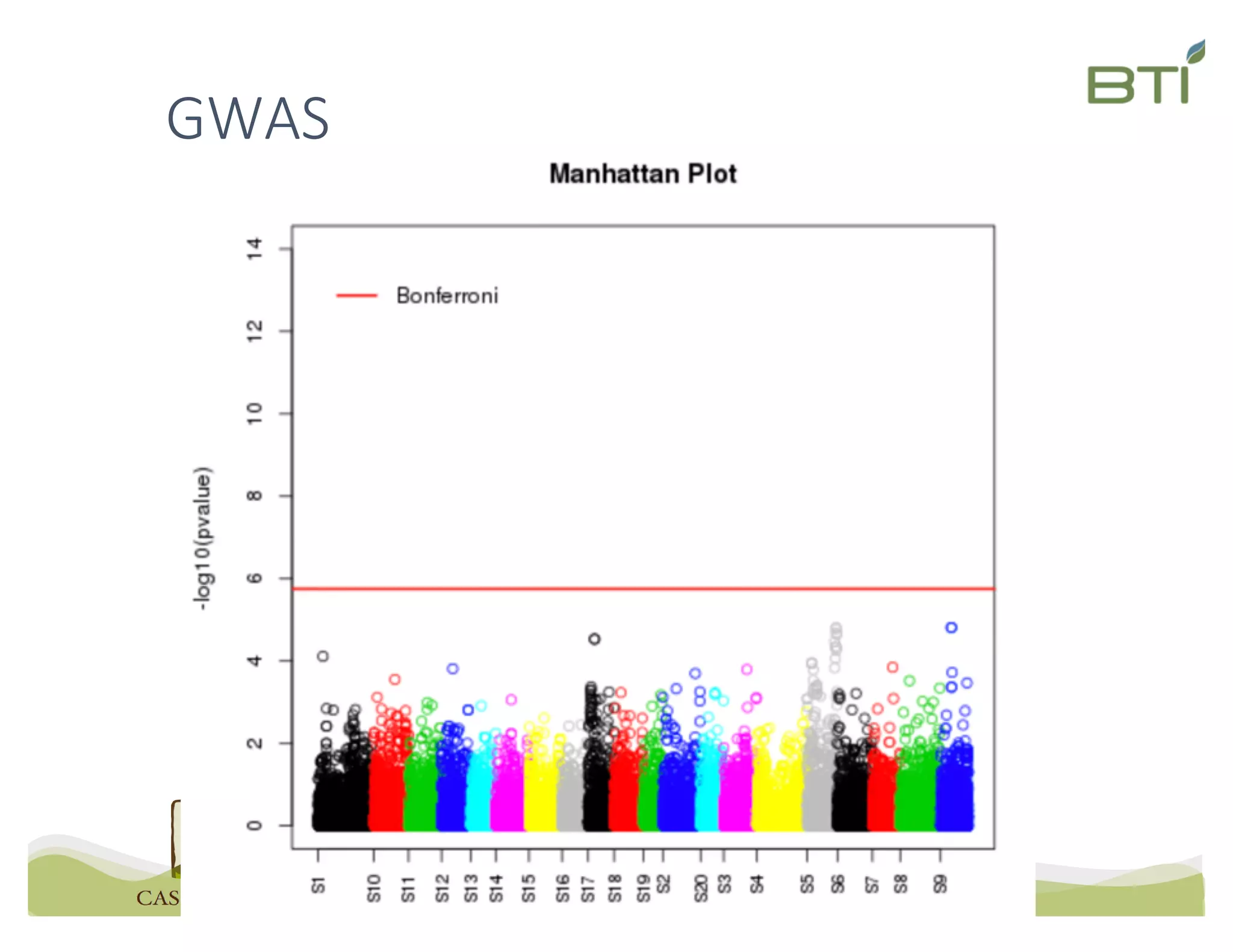
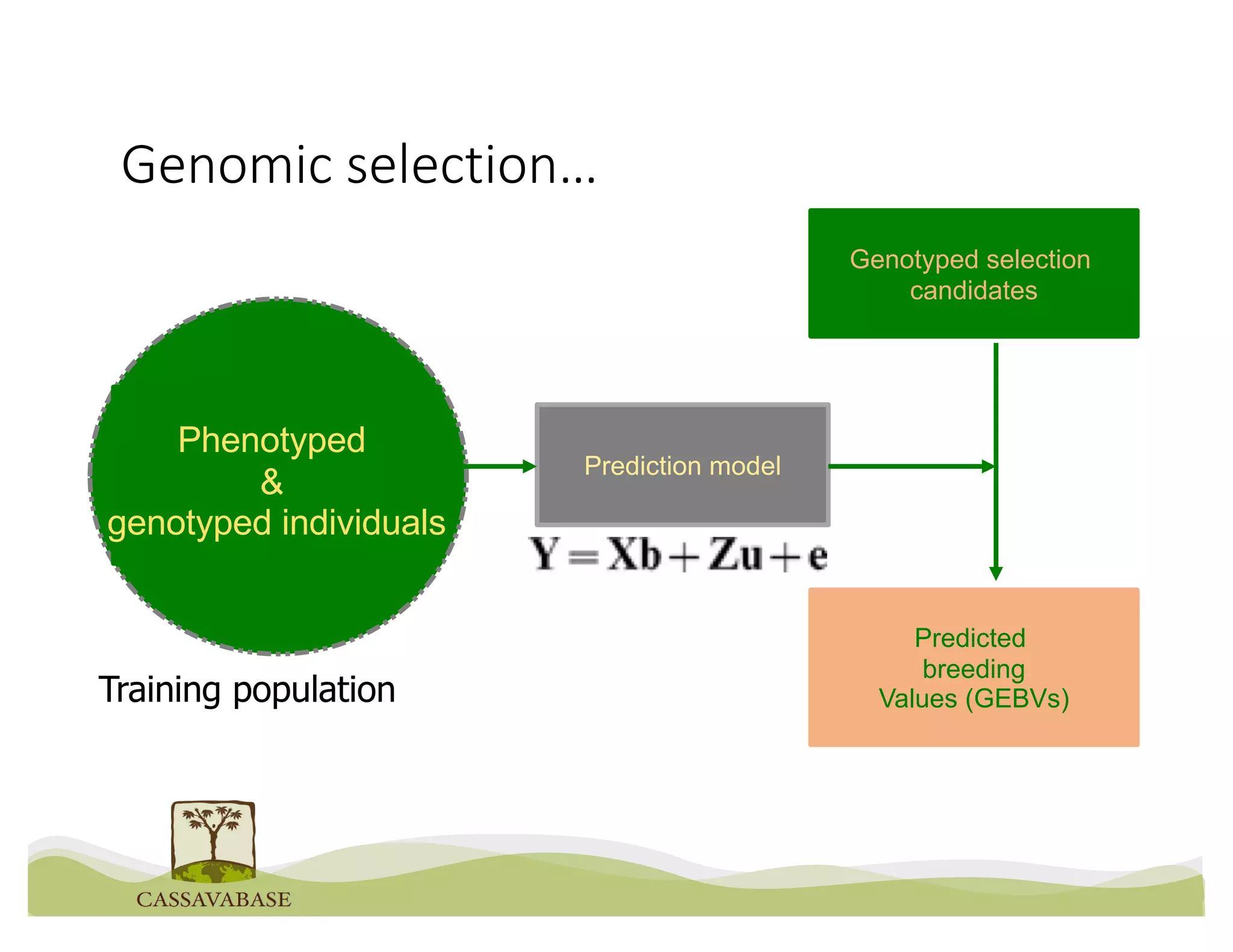
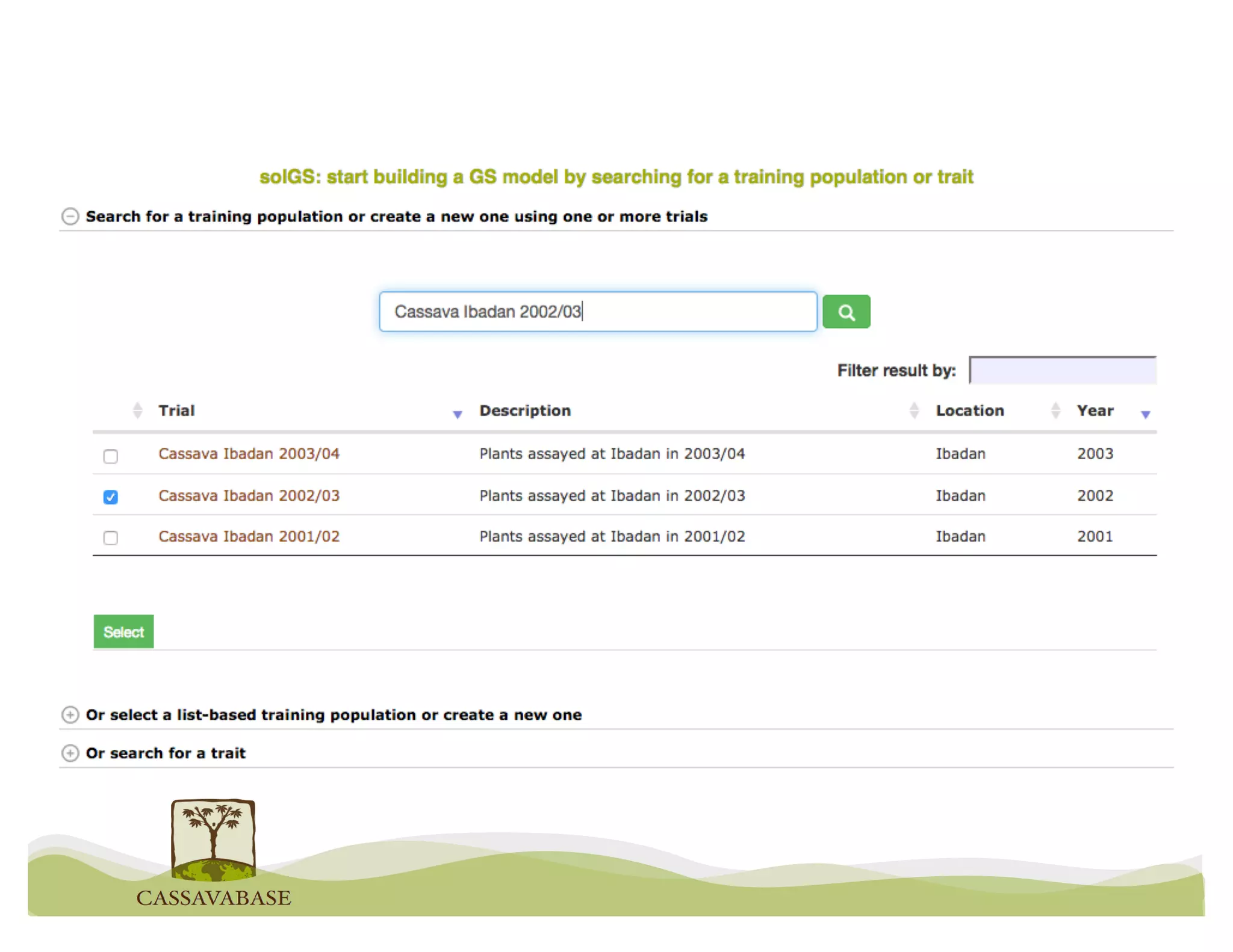
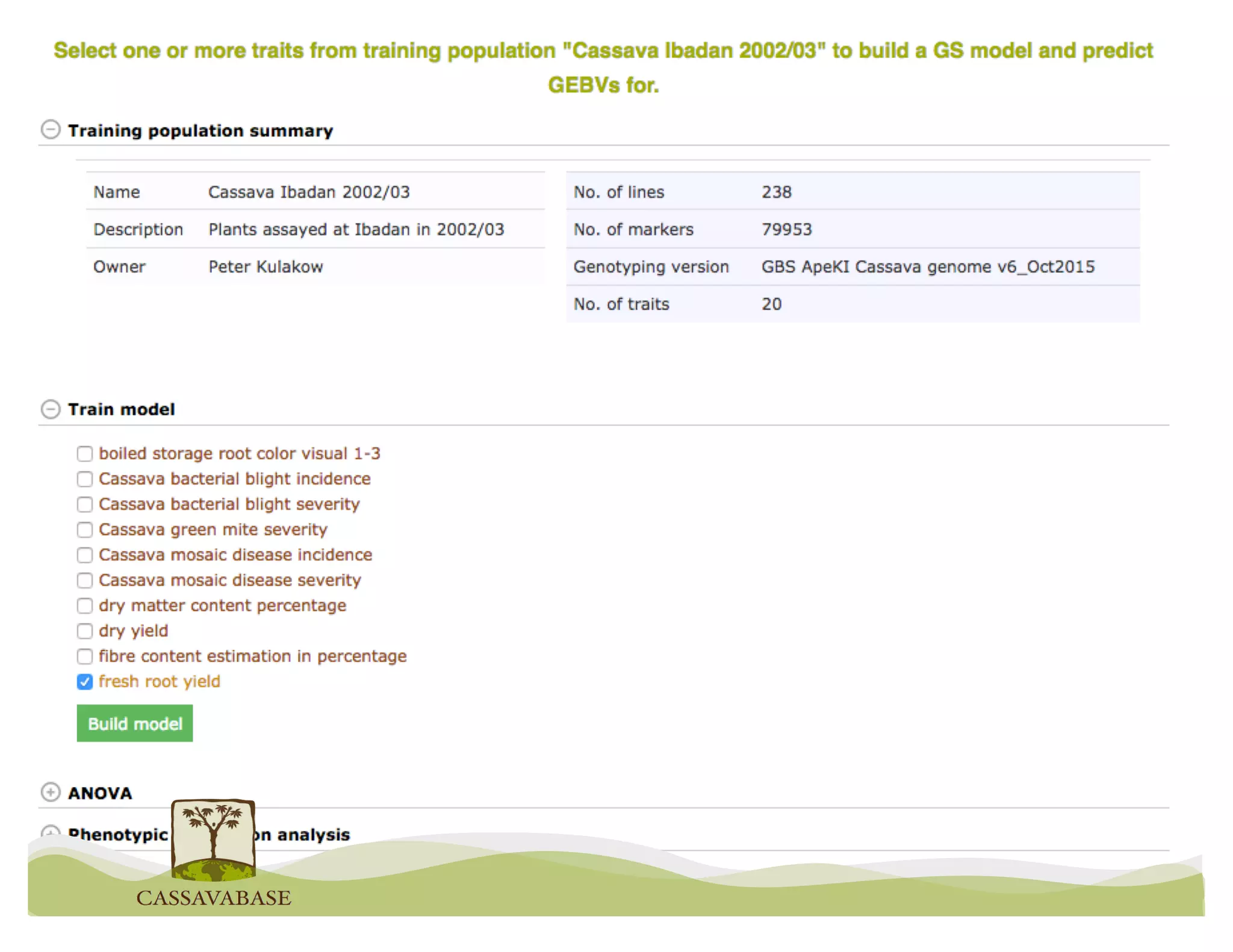
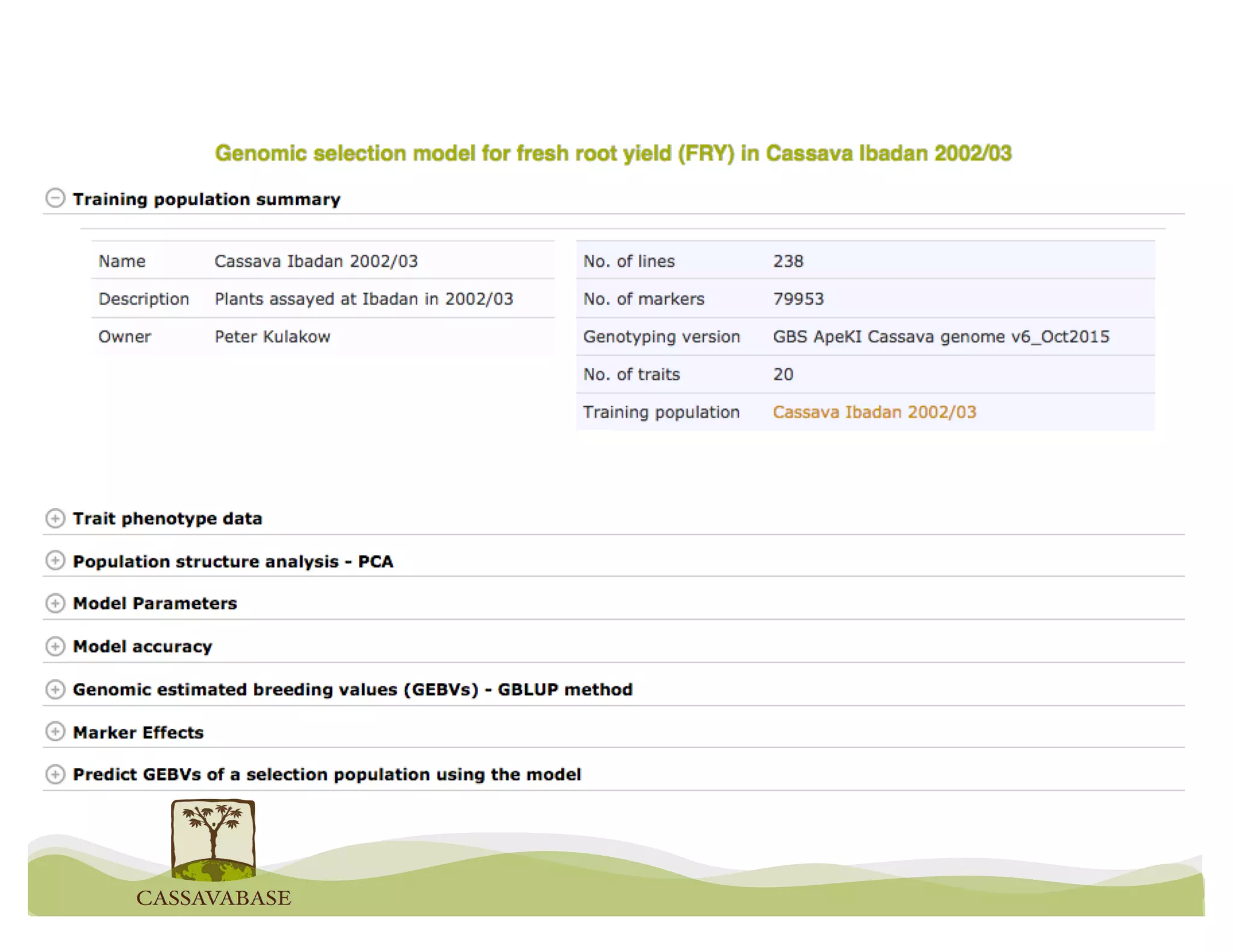
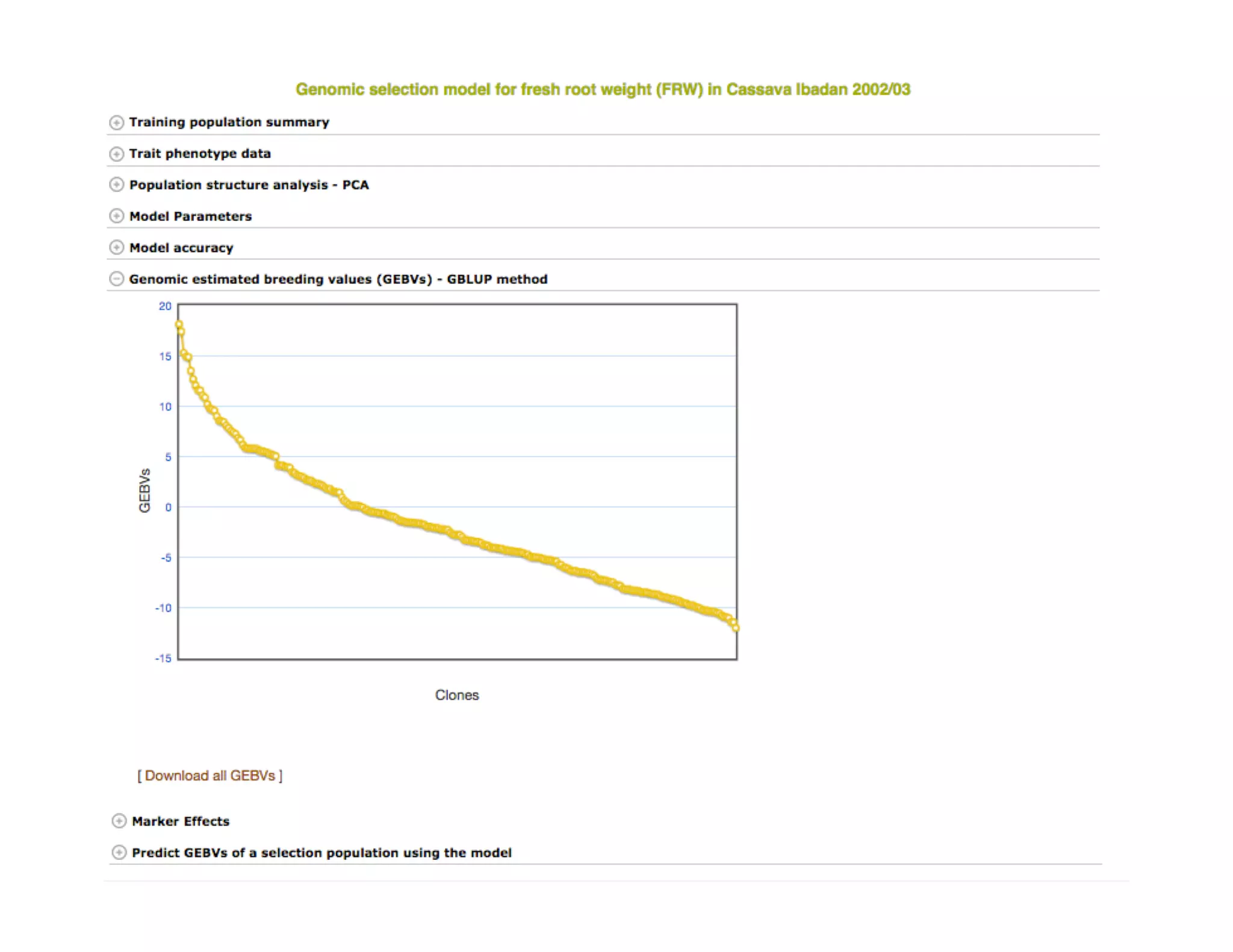
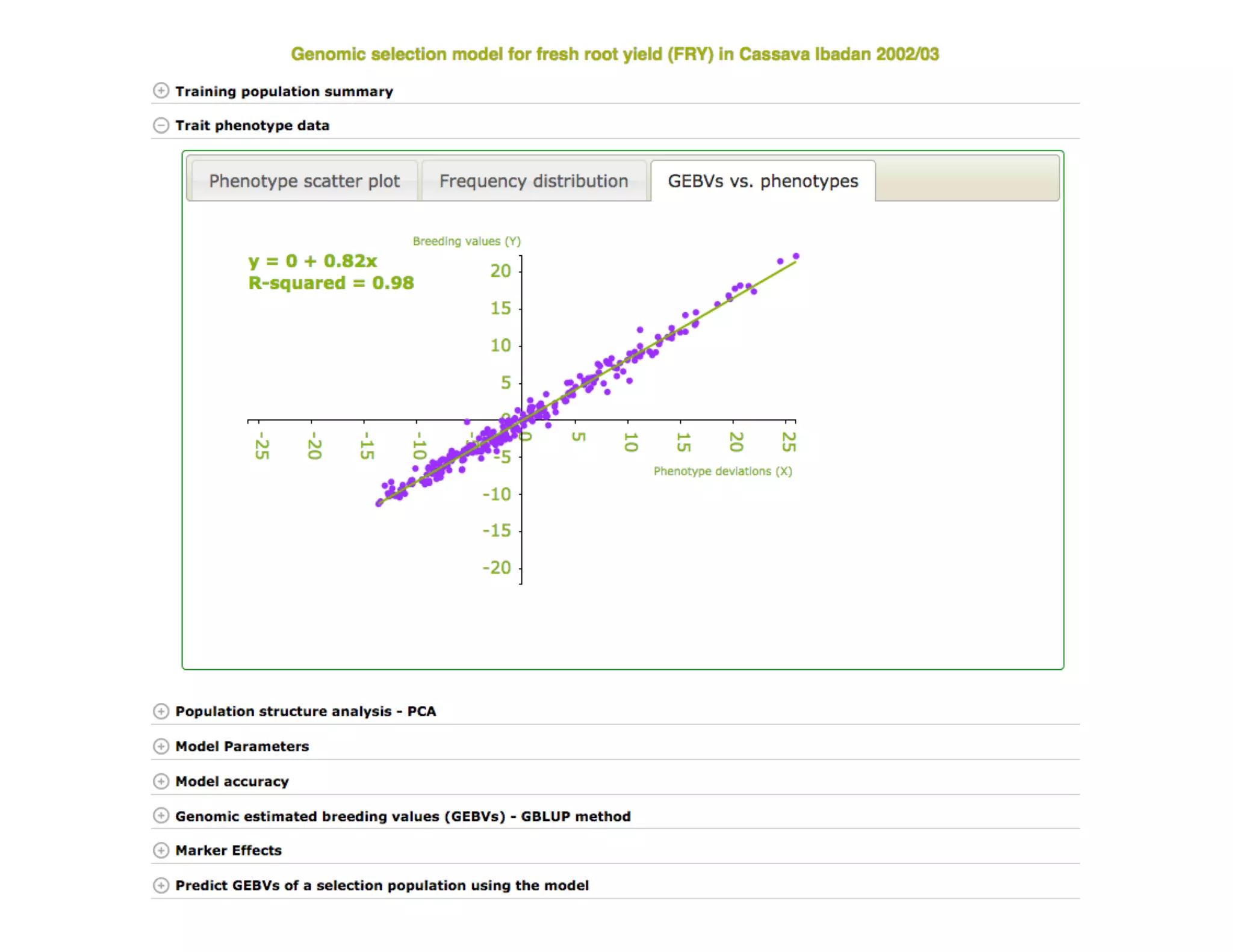
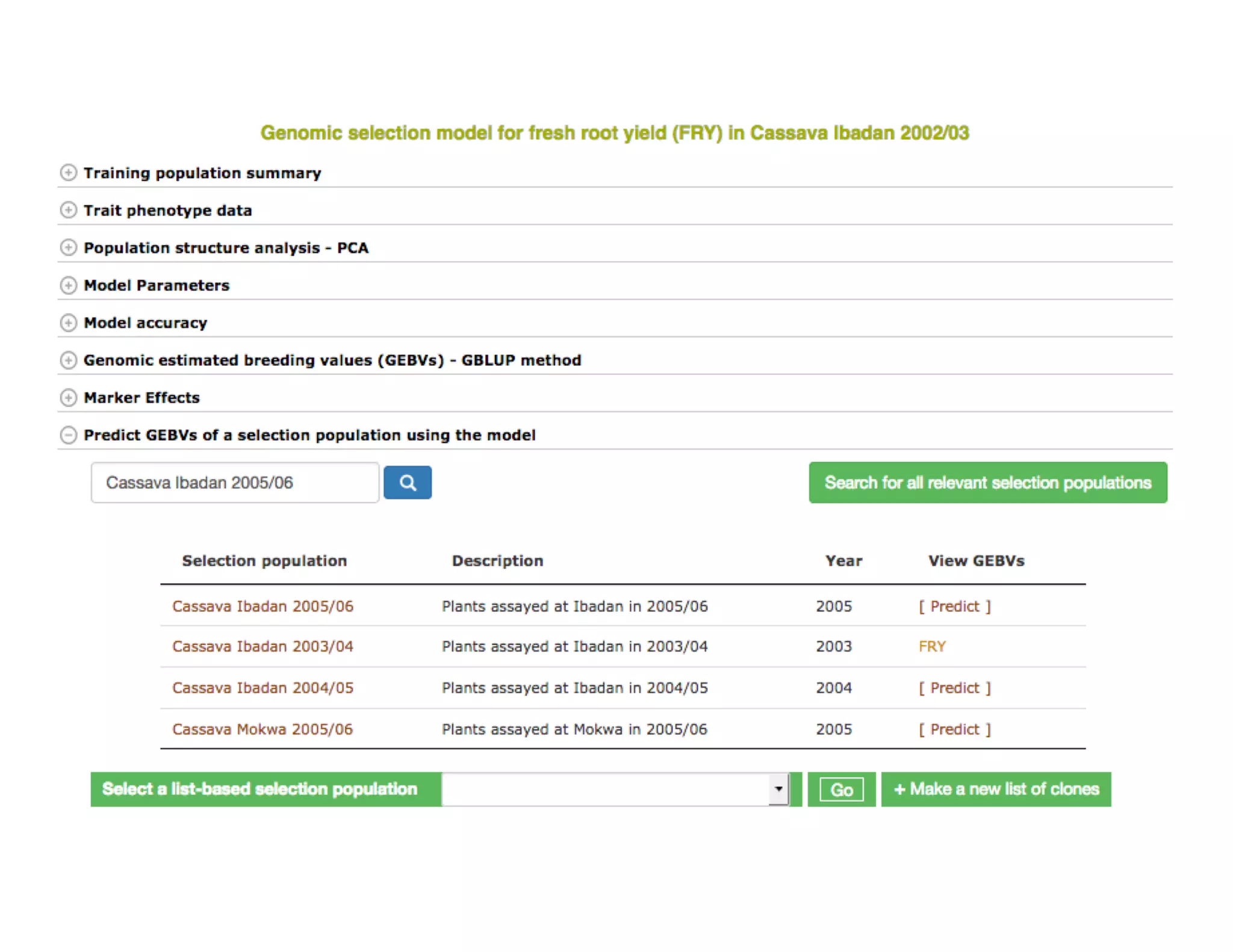
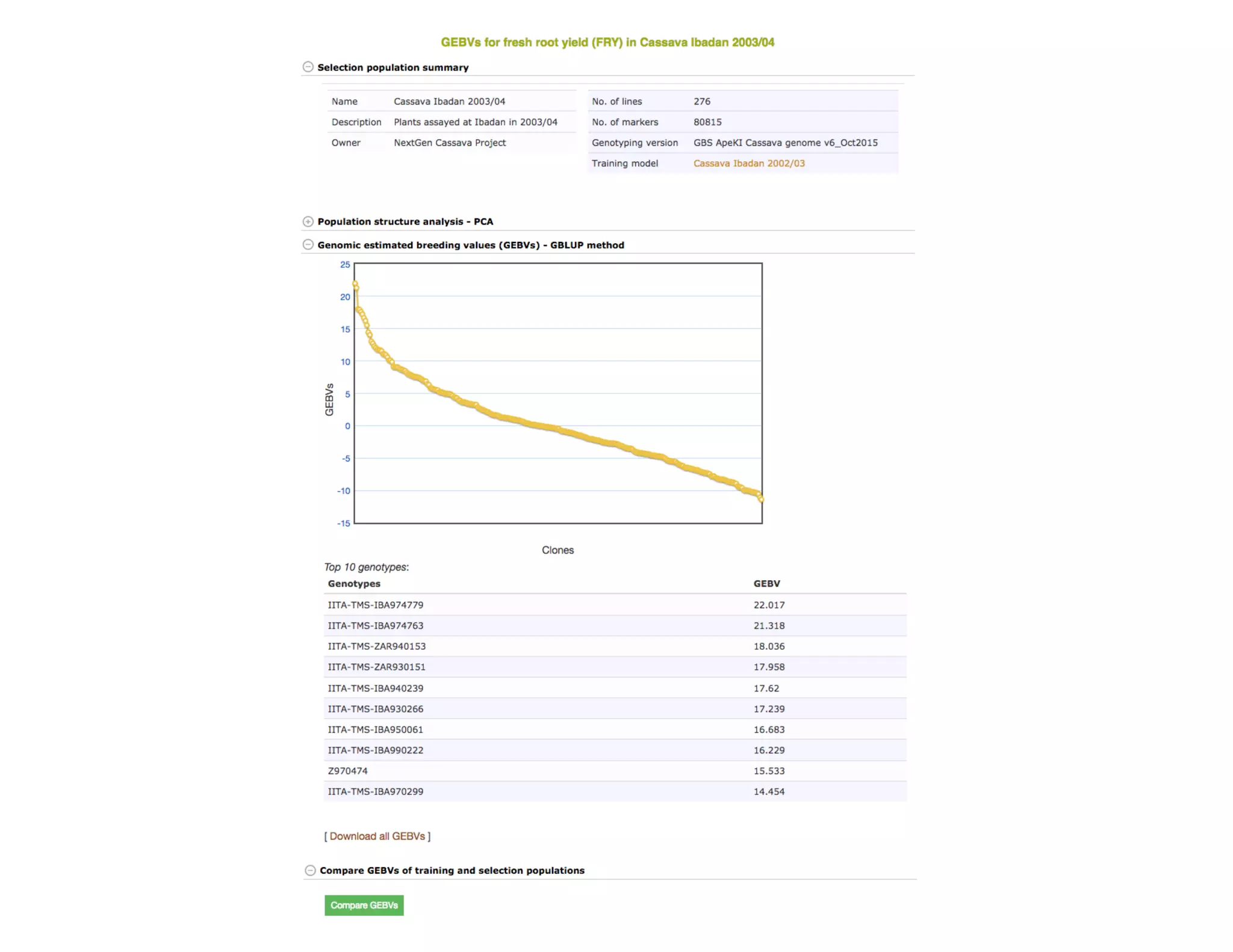
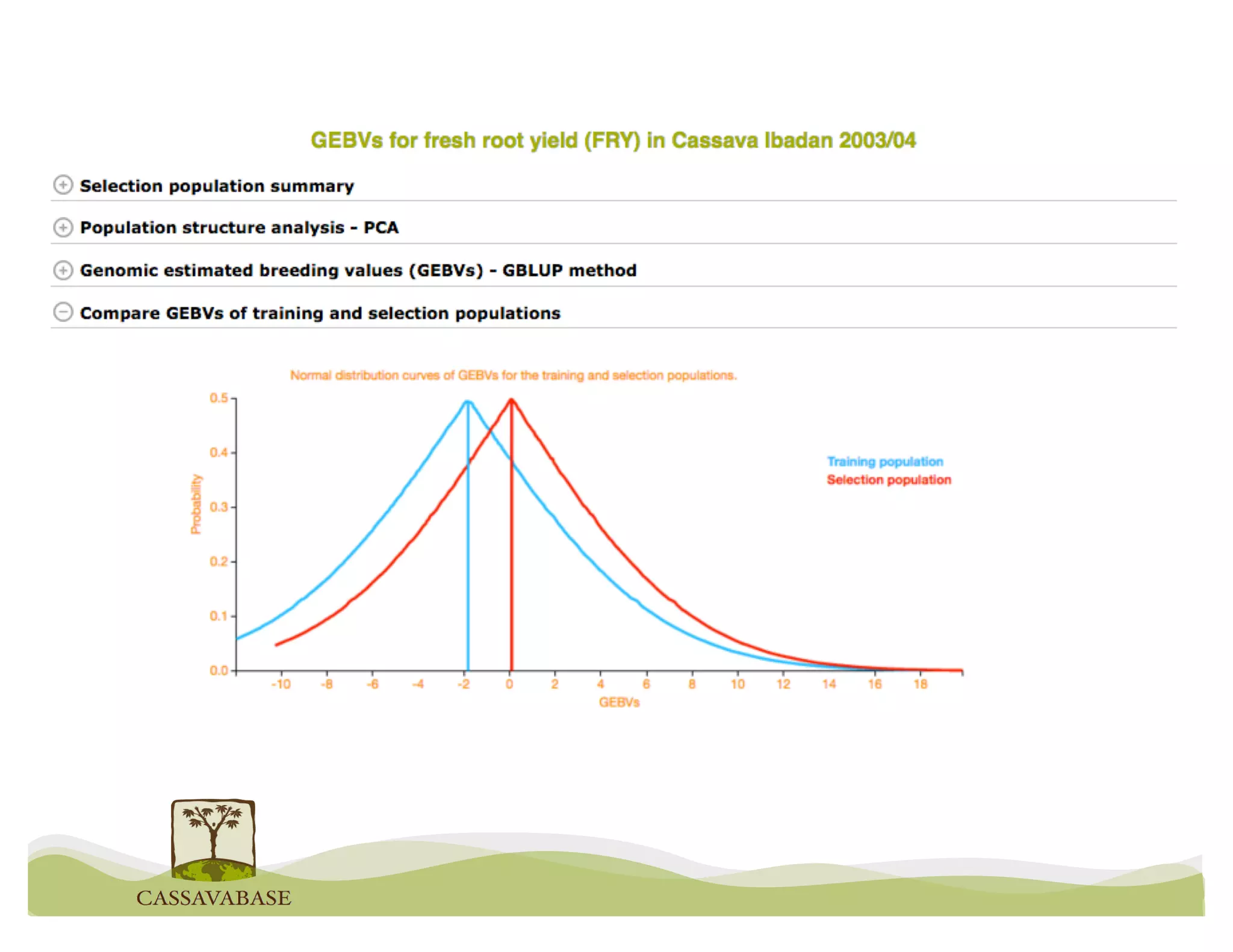
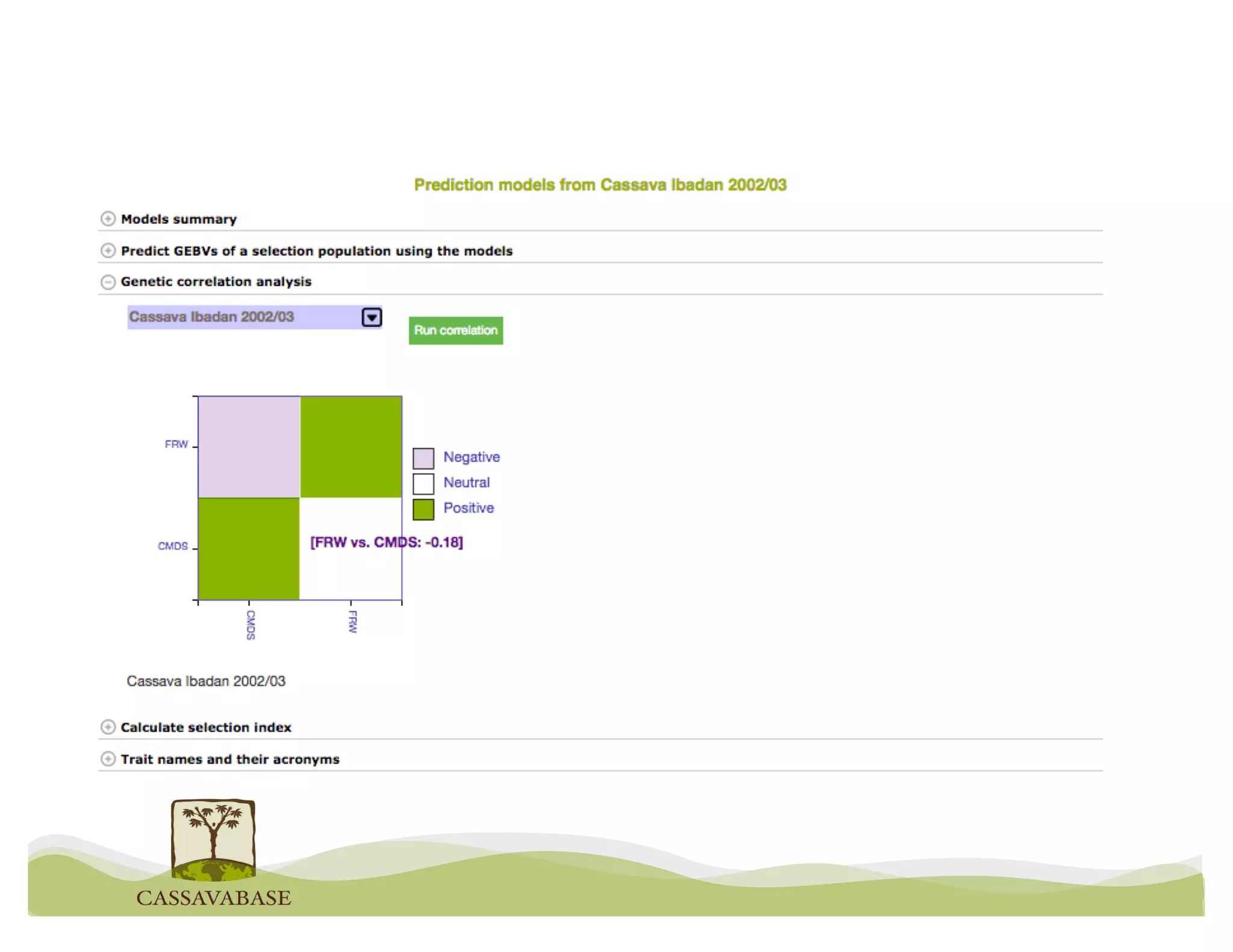
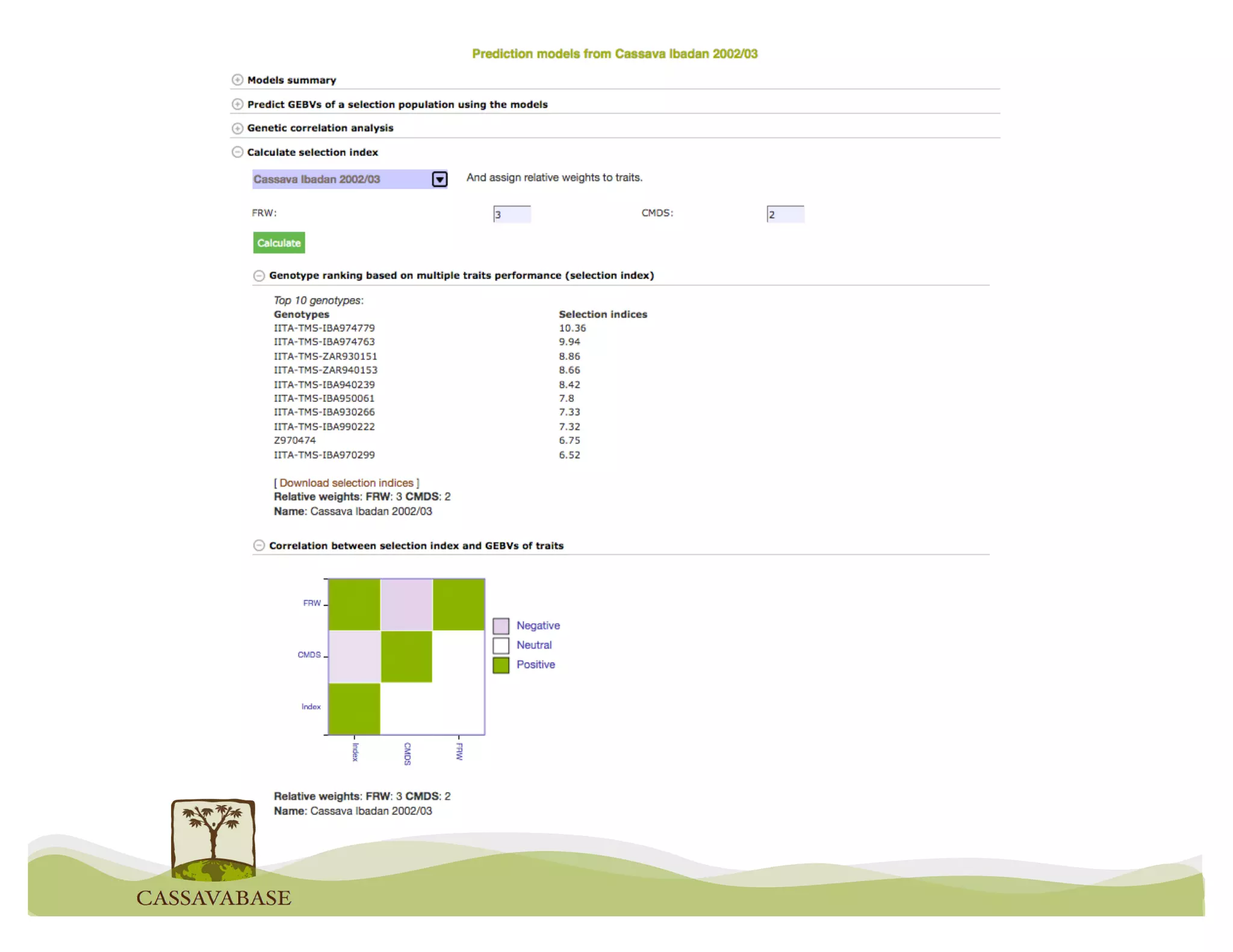
This document summarizes a presentation on breeding data management and analysis tools using the Cassavabase database. It describes how Cassavabase collects phenotypic and genotypic data from various programs and ensures data quality. Over 9 million phenotypic observations, 2488 trials and 34,000 genotypes have been collected. It also discusses expanding interoperability through APIs and partnerships with other crop databases. The presentation demonstrates using Cassavabase to access trial data, visualize genotypes, perform statistical analysis, genomic prediction and multi-trait selection to increase genetic gain.