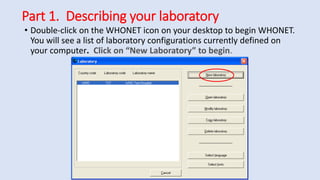
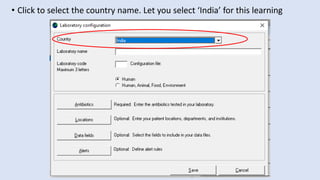
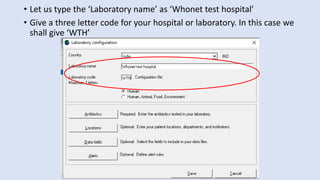
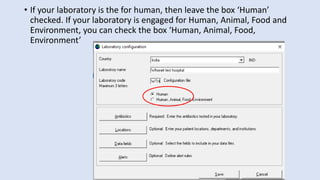
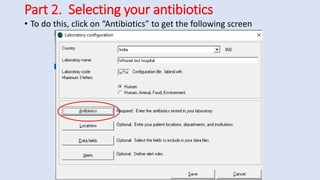
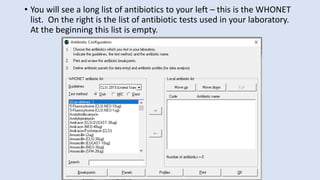
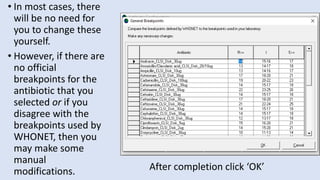
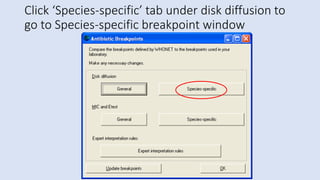
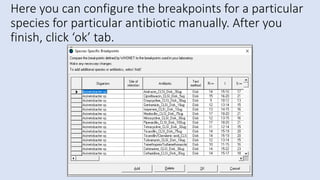
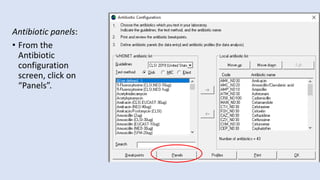
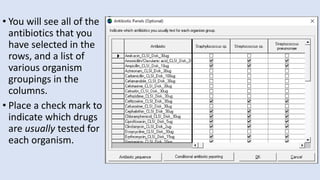
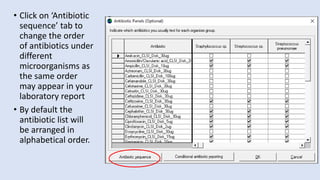
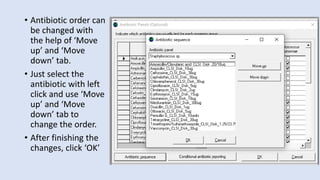
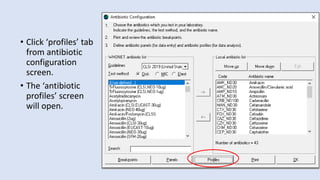
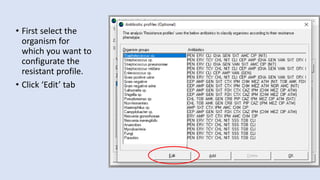
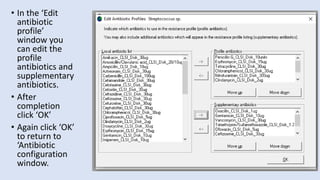
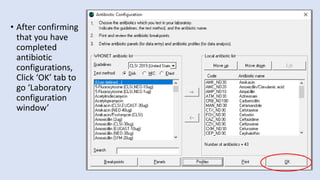
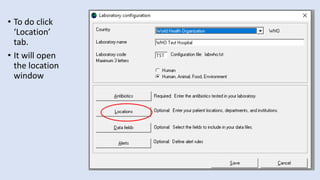
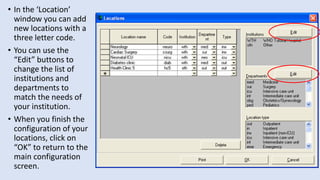
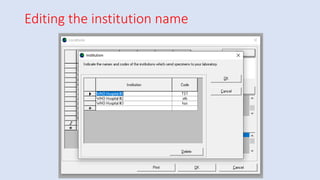
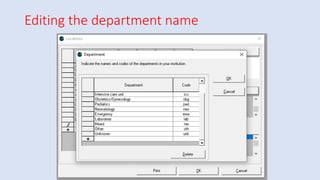
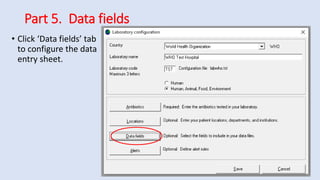
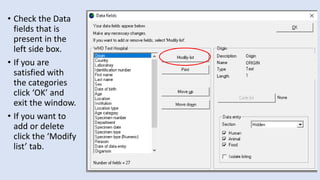
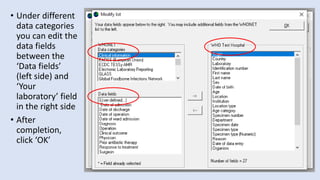
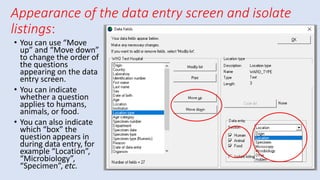
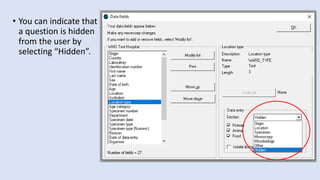
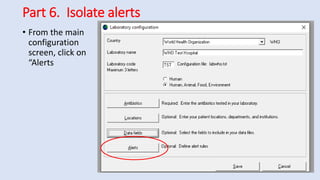
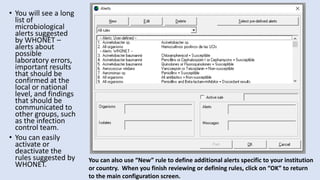
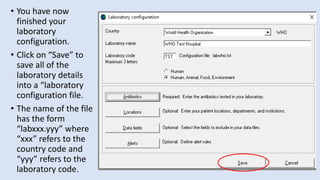
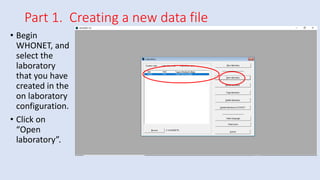
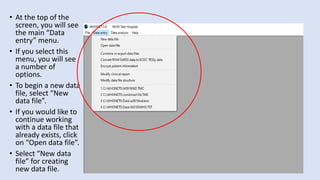
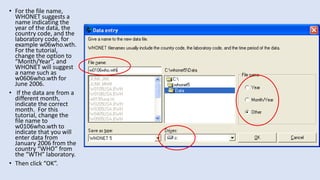
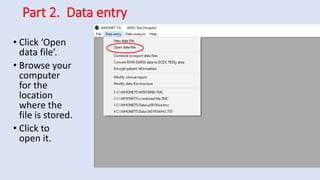
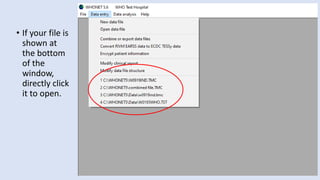
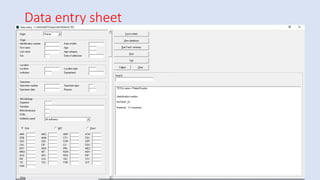
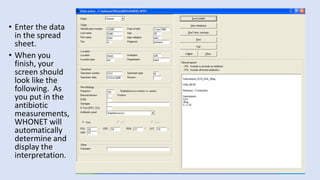
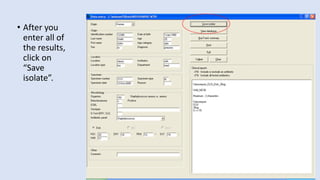
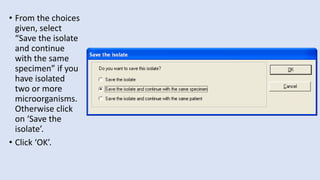
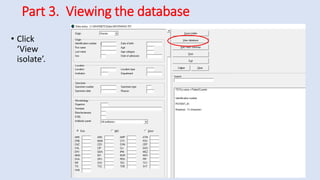
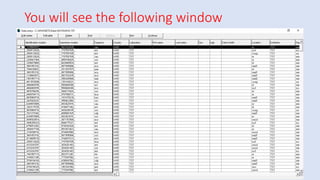
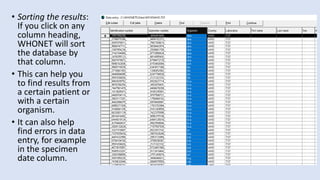
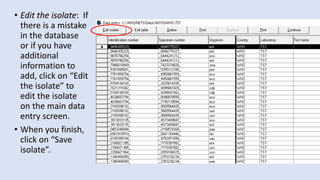
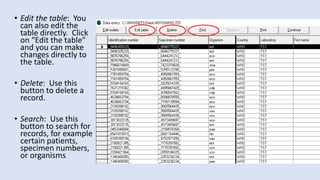
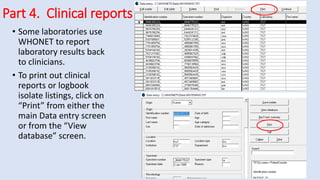
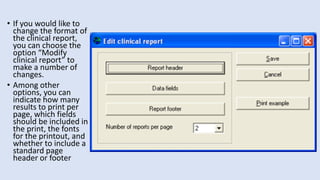
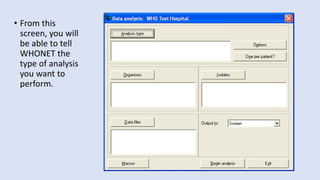
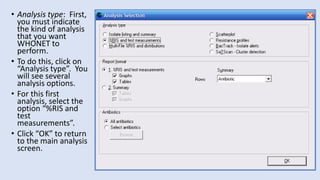
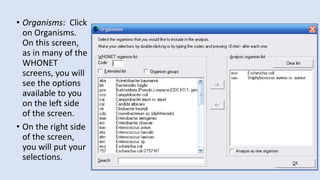
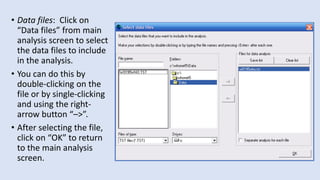
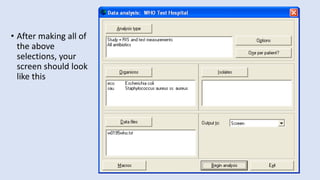
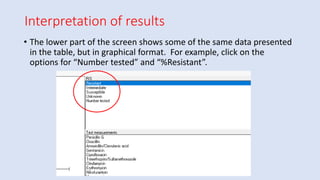
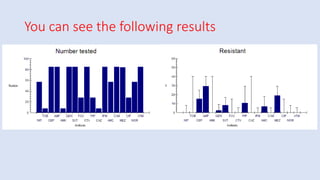
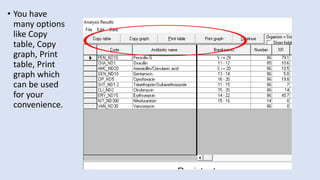
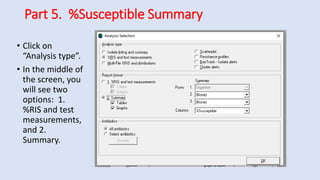
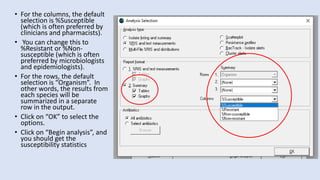
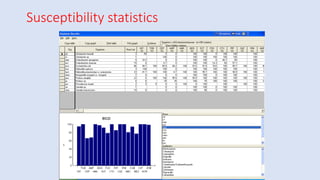
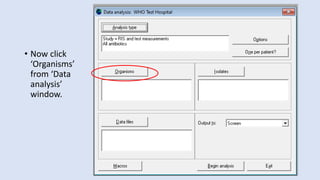
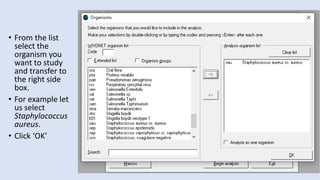
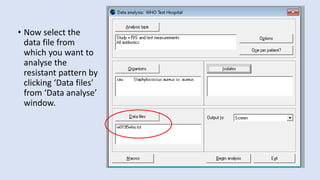
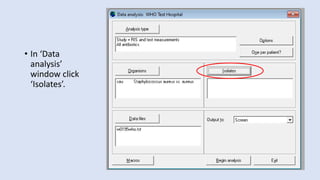
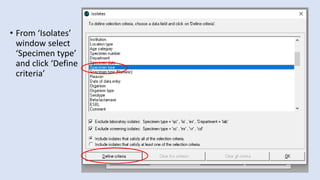
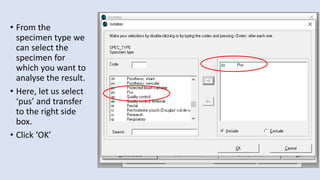
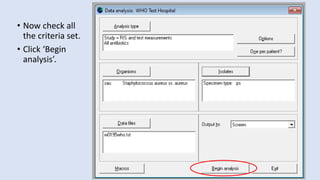
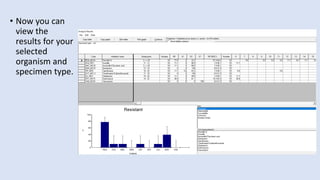
Whonet is a free software designed for the laboratory surveillance of infectious diseases and antimicrobial resistance, aiming to enhance local laboratory data use and facilitate collaboration. It features three main components: laboratory configuration, data entry and clinical reporting, and data analysis, allowing for customizable settings and detailed analysis tools for microbial populations. Whonet supports data management for bacteria, fungi, and parasites, aiding in the identification of outbreaks and quality assurance in laboratory testing.