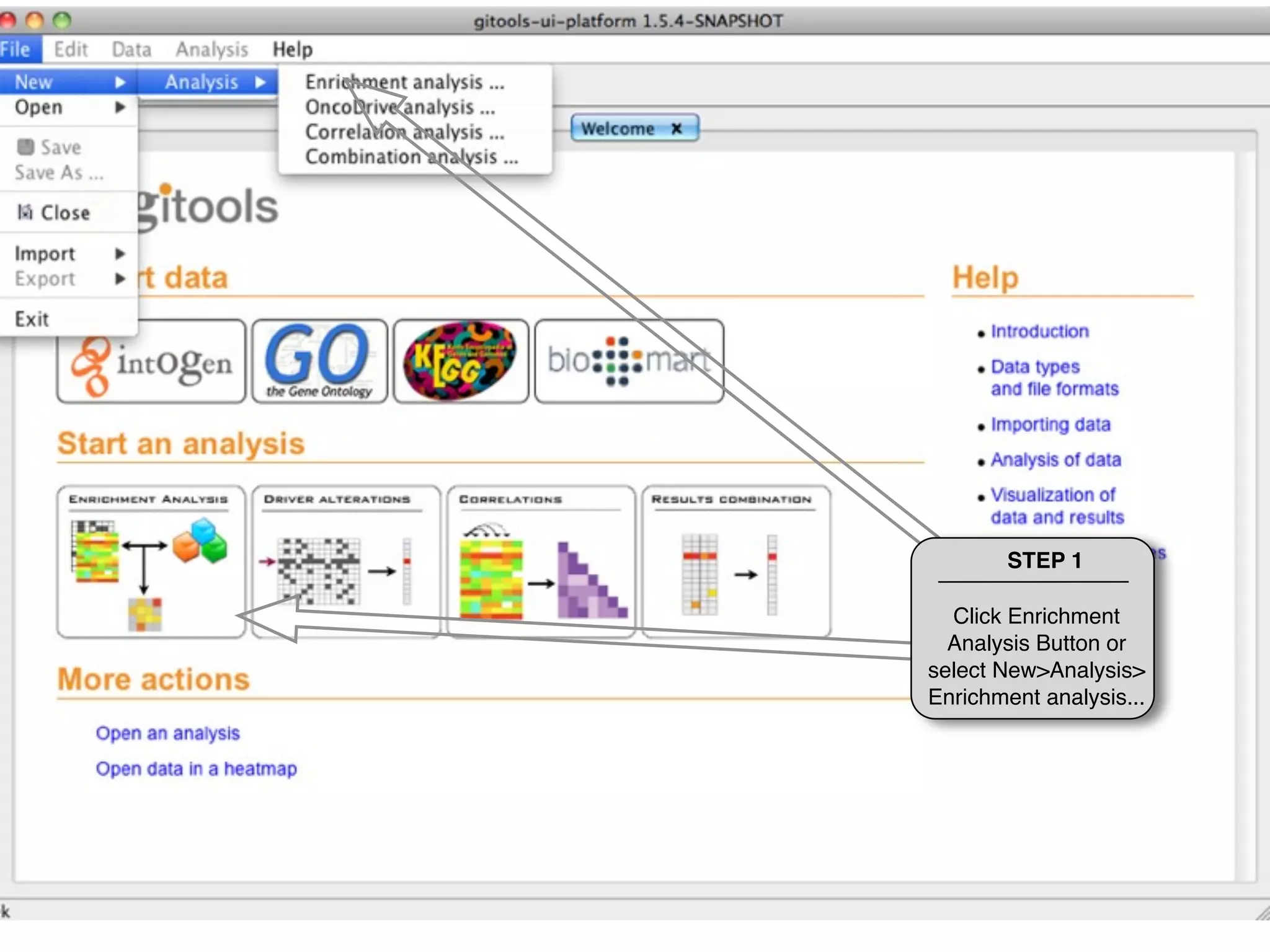
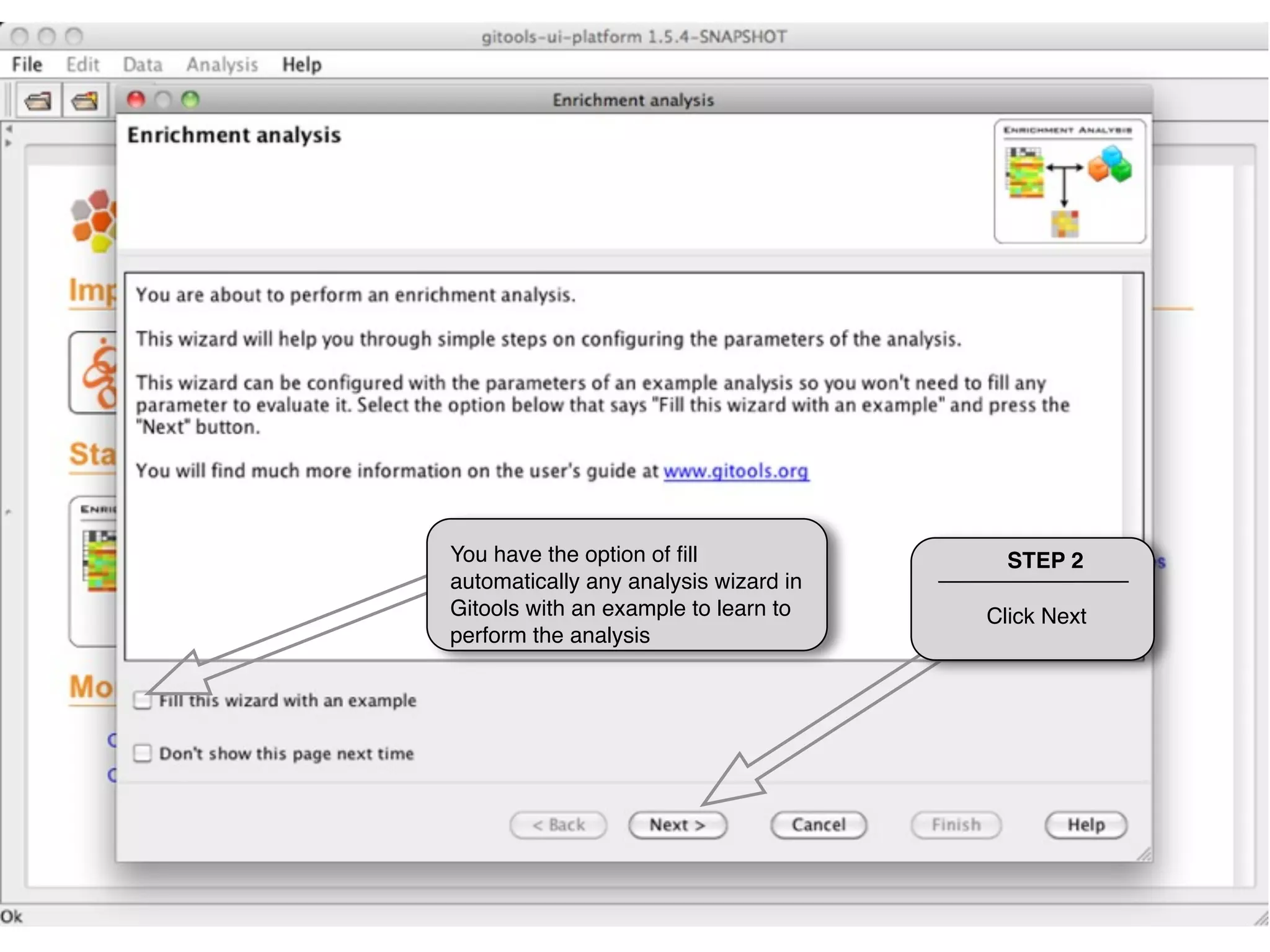
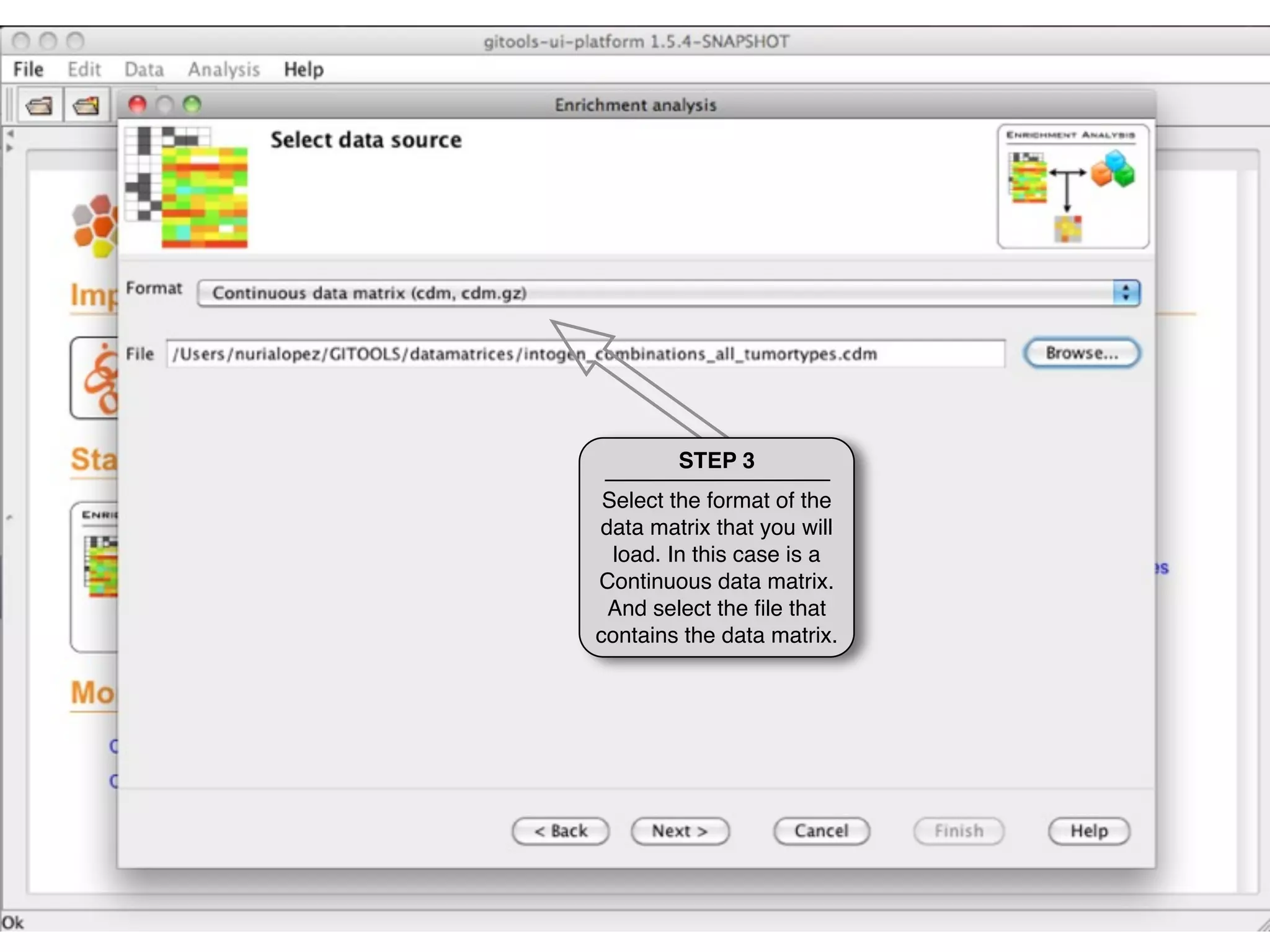
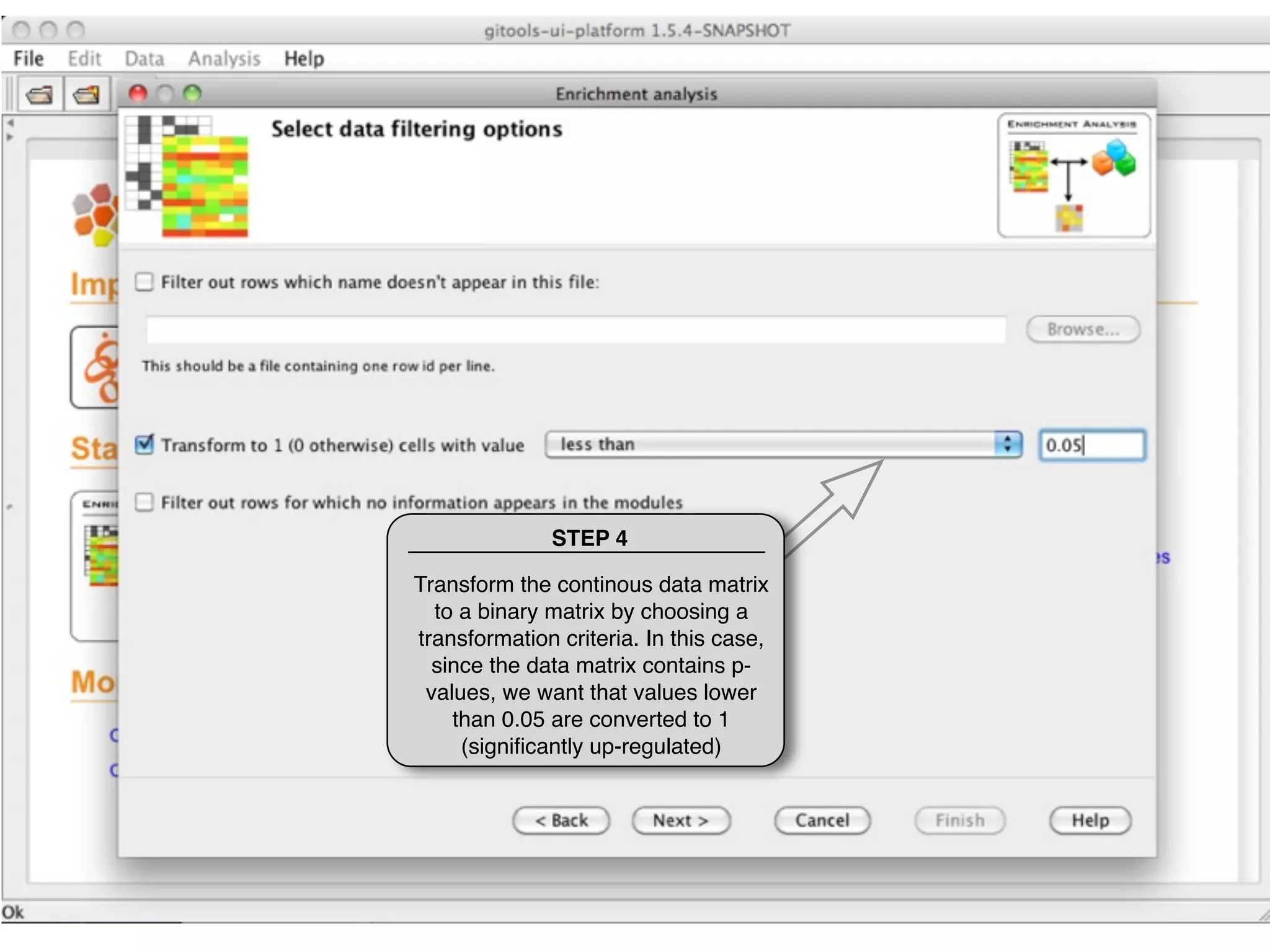
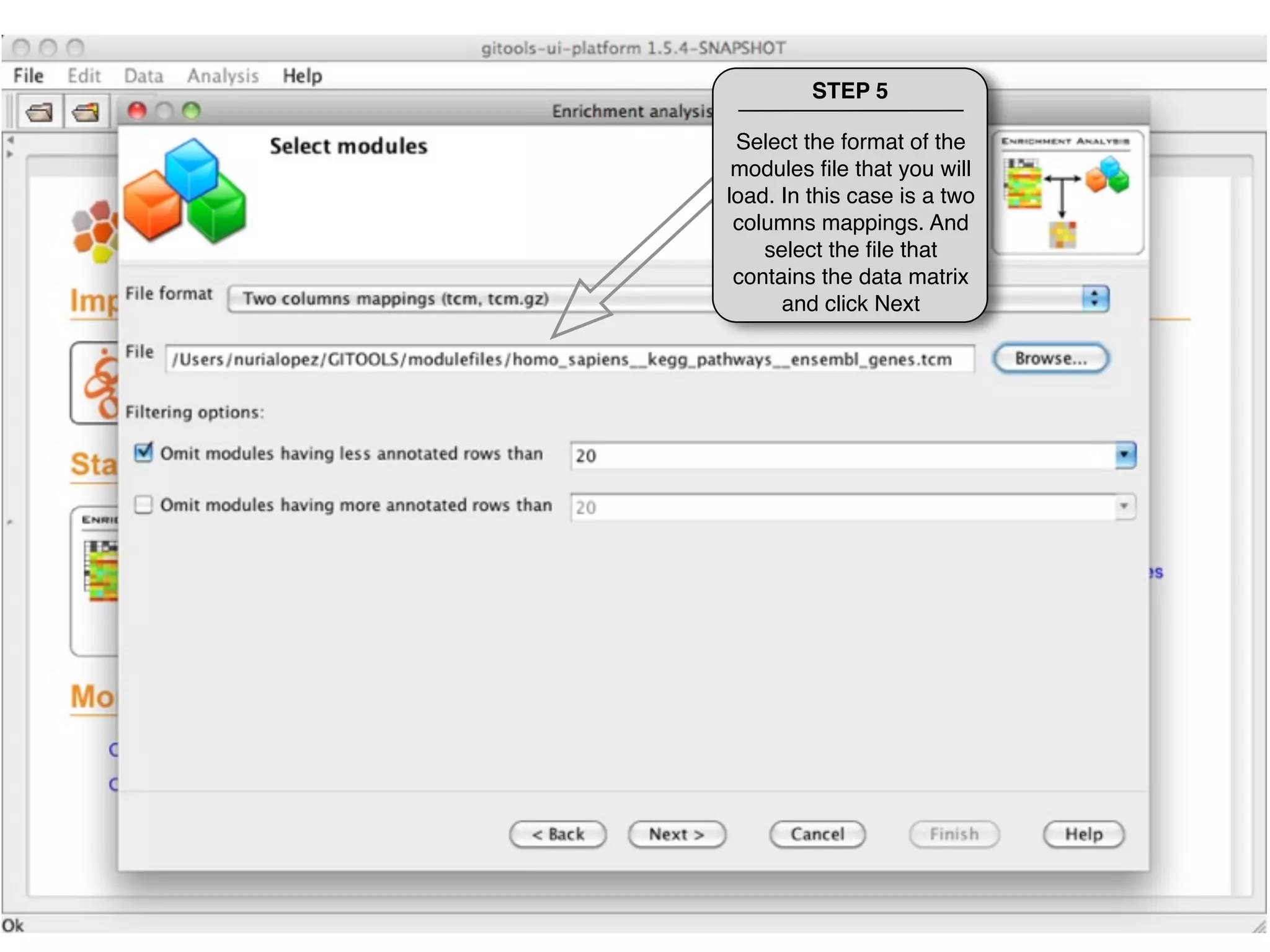
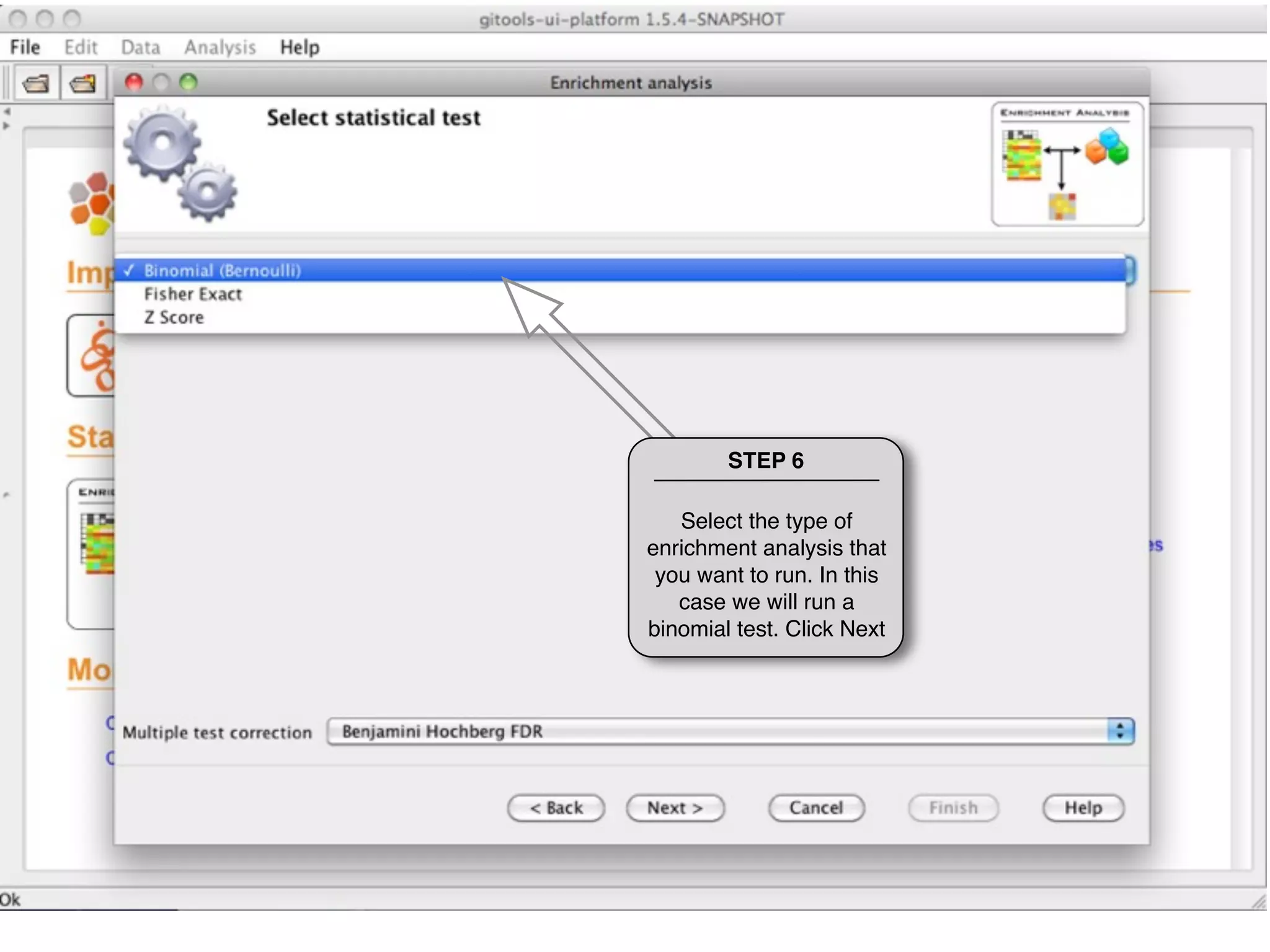
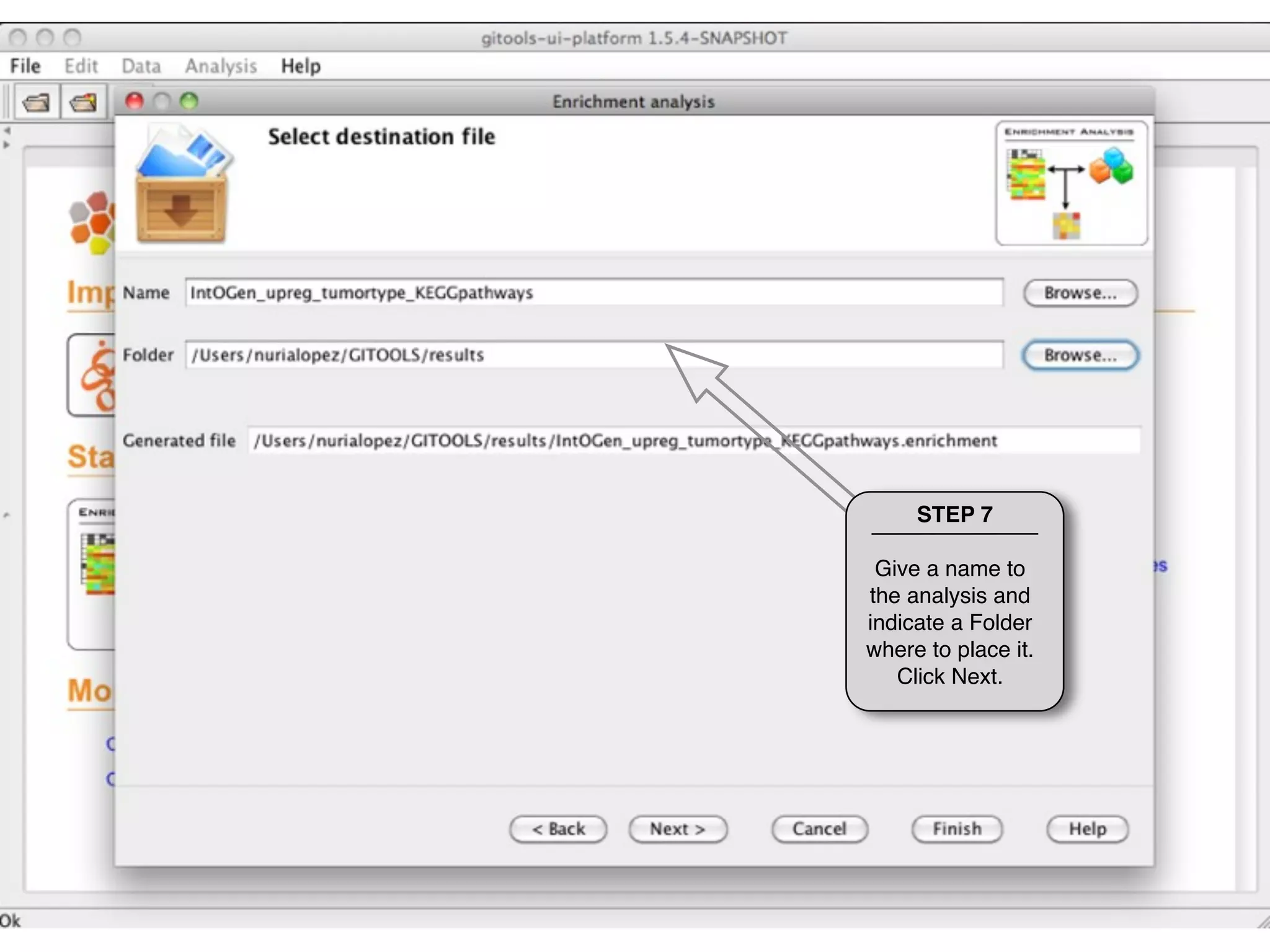
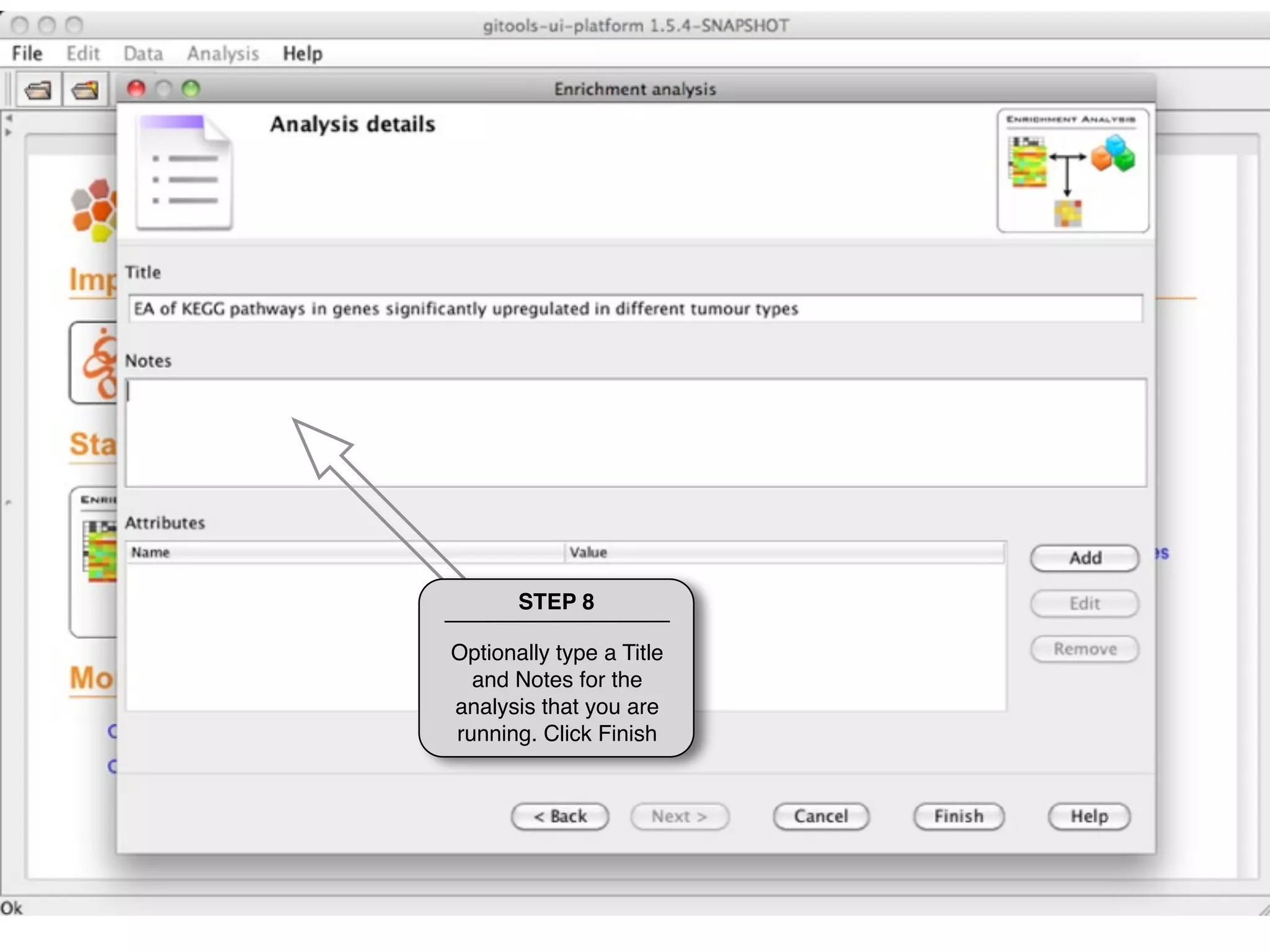
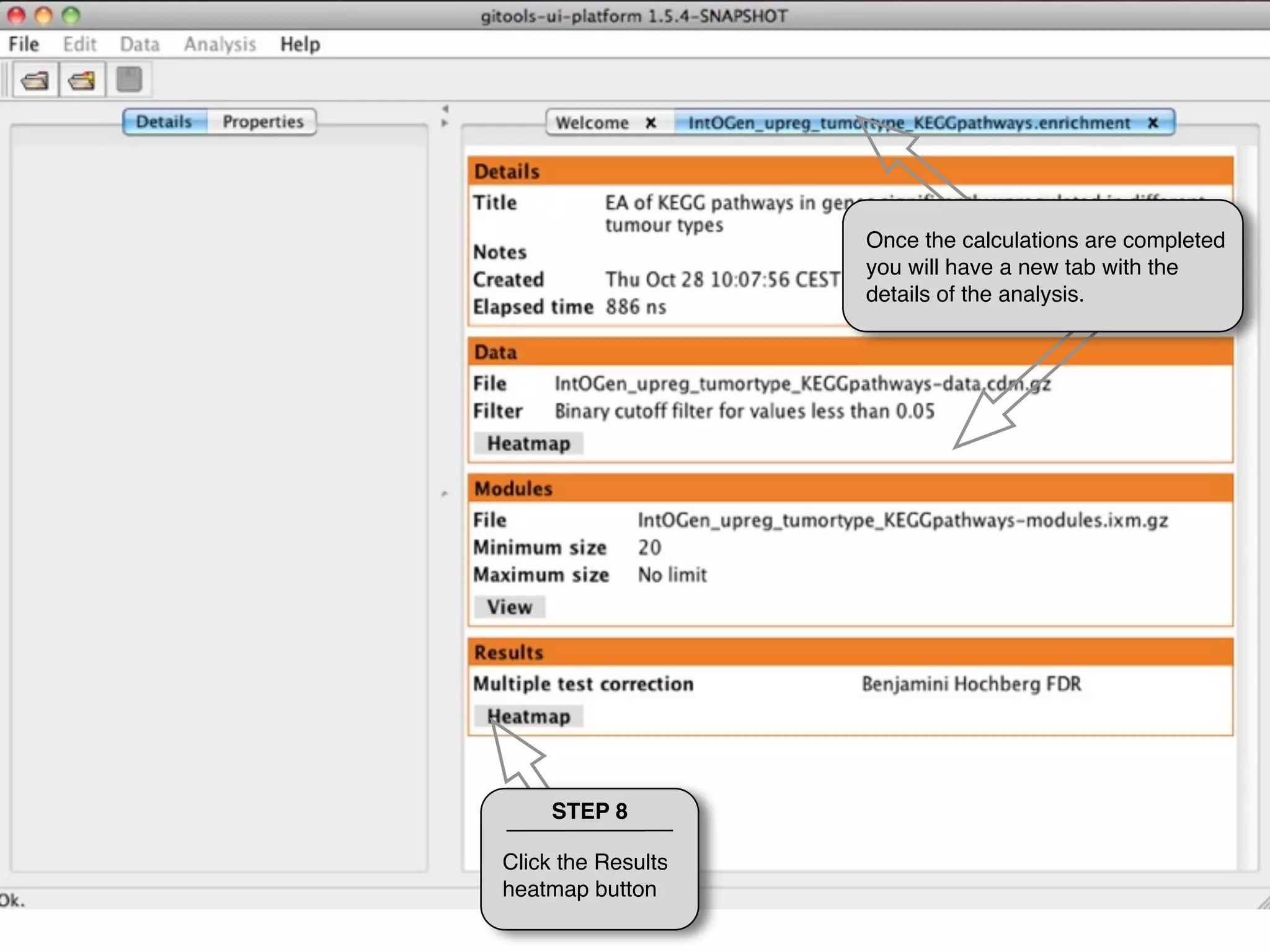
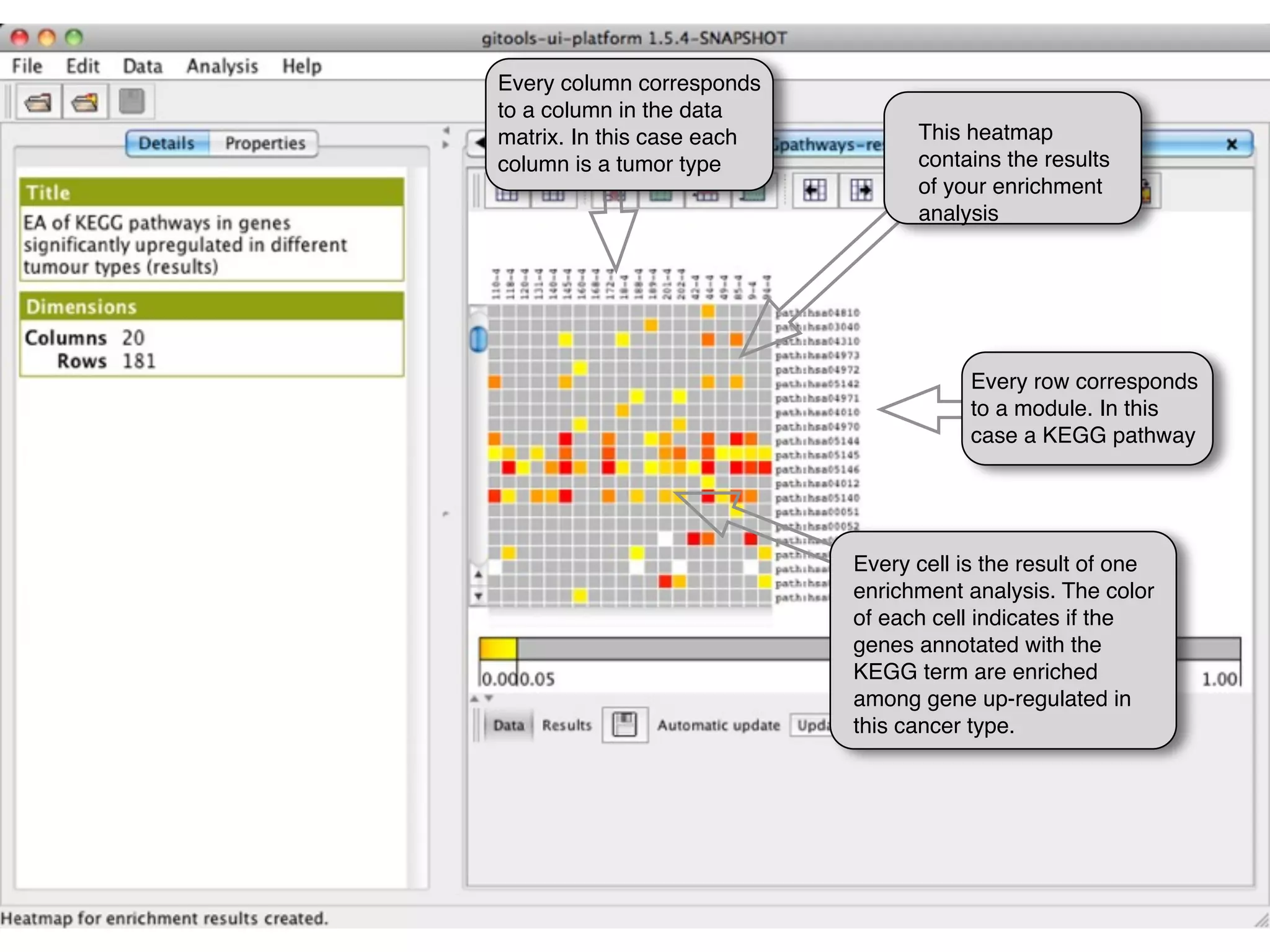
This document provides steps to run an enrichment analysis in Gitools to test which KEGG pathways are enriched among genes significantly up-regulated in different cancer types. The steps include: 1) selecting enrichment analysis, 2) loading a continuous data matrix and selecting significantly up-regulated genes, 3) running a binomial test, 4) naming and saving the analysis, and 5) viewing results in a heatmap where columns are cancer types and rows are enriched KEGG pathways.