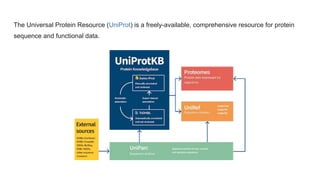
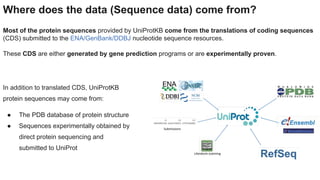
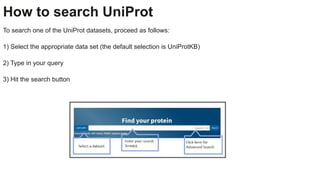
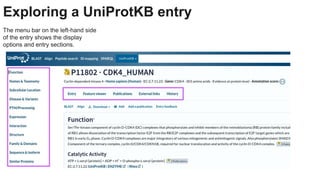
This document provides an overview of the Universal Protein Resource (UniProt) database. It discusses that UniProt is a comprehensive resource for protein sequence and functional data. Most protein sequences in UniProt come from gene predictions or experimental evidence. The document outlines how to search and explore UniProt entries, including how annotation scores are determined and what different evidence levels indicate. It provides guided examples of using UniProt to find protein function, link a disease to a protein and variant, and download a proteome set.