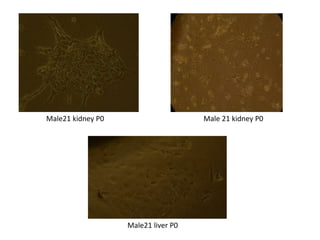
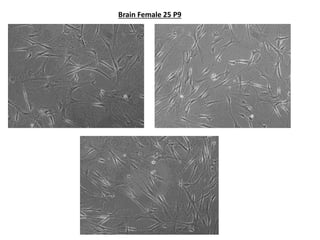
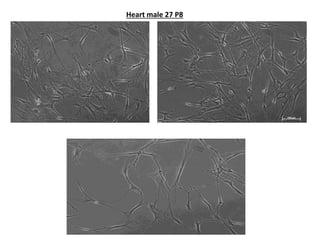
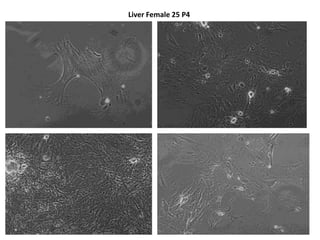
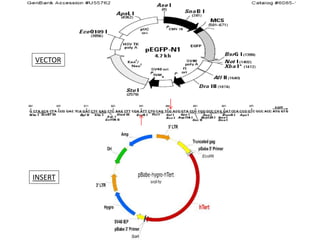
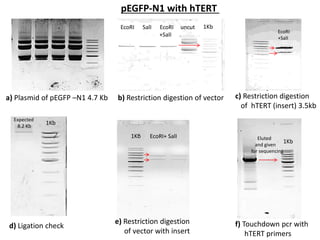
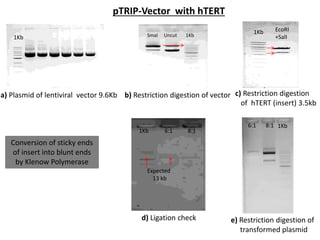
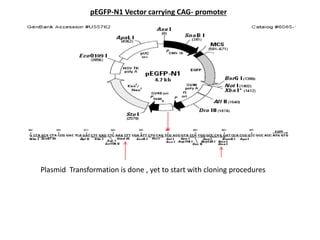
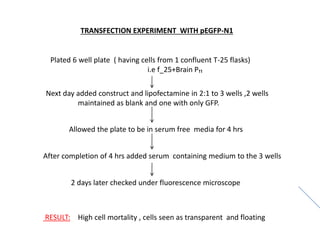
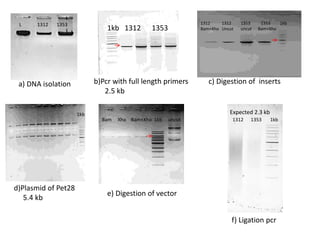
This document summarizes work done on culturing crocodile cell lines and cloning the parc gene from Pseudomonas keratitis. Primary crocodile cell lines were established from various organs and immortalized using hTERT. The parc gene was cloned from mutant and wild-type Pseudomonas strains and will be expressed and crystallized to study its role in quinolone antibiotic resistance.