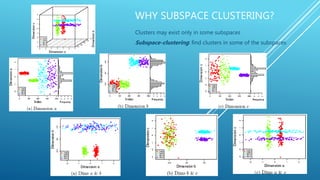
This document discusses the challenges of clustering high-dimensional data due to the curse of dimensionality, leading to the exploration of subspace clustering methods that identify clusters in specific dimensions. Two notable algorithms are presented: Clique, which utilizes density and grid-based approaches to partition data into dense units, and Proclus, which defines clusters and performs a systematic process for medoid selection and outlier detection. The overall goal is to enhance clustering effectiveness by focusing on relevant subspaces rather than relying on a uniform distance measure across all dimensions.