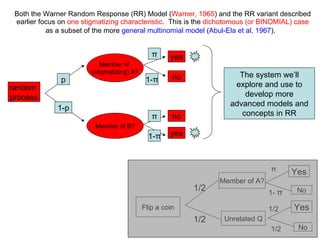
This document discusses simulations of multinomial randomized response models. It describes how the Warner randomized response model was extended to allow for multiple mutually exclusive categories. The key points are:
1) Abul-Ela et al. (1967) defined the multinomial randomized response model which allows sampling of multiple groups during a single experiment.
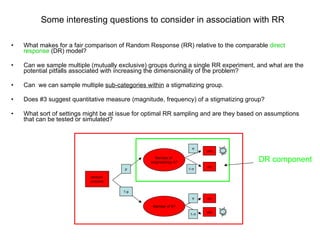
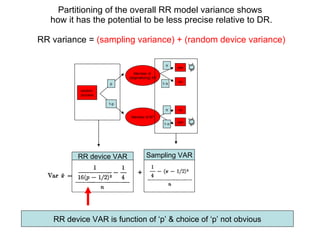
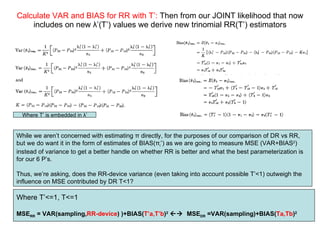
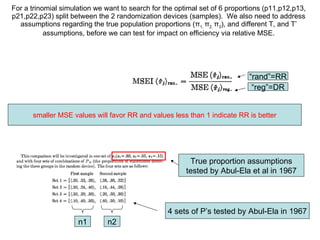
2) Simulations of the trinomial randomized response model are discussed, including calculating variances, biases, and mean squared errors to compare to direct response models.
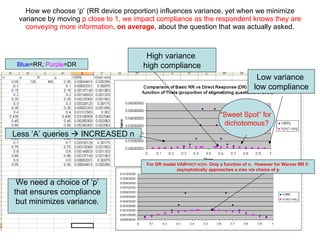
3) Optimal values of the randomization probabilities (p-values) need to be determined to minimize variance while maximizing compliance for the trinomial model.


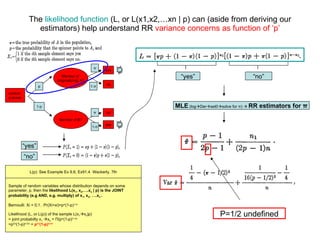
![Likelihood (L) shows that VAR of the RR device & compliance (or truth) diametrically opposed as a function of ‘p’ . L(p, π ) = [ π p+(1- π )(1-p)] n1 [(1- π )p+ π (1-p)] n-n1 L( p=1, π ) = [ π +(1- π )(1-1)] n1 [(1- π )+ π (1-1)] n-n1 L( π ) = [ π )] n1 [1- π ] n-n1 If p=1 : L(p, π ) L( π ), in effect it becomes a DR model where likelihood depends entirely on π , therefore respondent no longer anonymous. L(p, π ) = [ π p+(1- π )(1-p)] n1 [(1- π )p+ π (1-p)] n-n1 L( p=1/2, π ) = [ π /2+(1- π )(1-1/2)] n1 [(1- π )1/2+ π (1-1/2)] n-n1 L(p=1/2, π ) = [ π /2+1/2- π /2)] n1 [1/2- π /2+ π /2] n-n1 L( p )=(1/2) n1 (1/2) n-n1 If p=1/2 L( p=1/2, π ) L( p ) likelihood does not depend on π , only depends on p , therefore no information is imparted by the sampling operation: e.g. nothing learned . The closer we get to p=½ (in this model) the less we know and the higher our variance, and the closer we get to p=1, the lower our variance but the less anonymity & compliance will be achieved.](https://image.slidesharecdn.com/rrtrh51-12706549963629-phpapp02/85/Multinomial-Model-Simulations-7-320.jpg)

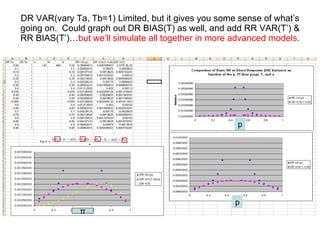
![Let’s re-parameterize the DR model further by the inclusion of T , the probability of telling the truth . T will impact DR VAR and DR BIAS. So is the whole story represented by a low DR variance [V( π , n )] relative to RR (device, p) added variance [V( π , n, p )] ?](https://image.slidesharecdn.com/rrtrh51-12706549963629-phpapp02/85/Multinomial-Model-Simulations-10-320.jpg)
![For a reasonable simulation w e need acknowledge known large magnitude DR BIAS, as well as additional variance based on compliance, as compared to the added RR device variance. T = probability of telling the truth, T (0 1), Ta=prob for group A, Tb=prob for group B 1) We can assume either that we have chosen a p that implies both Ta=Tb = 1 in the RR settings, that is, a ‘p’ that elicits the TRUTH , in that we will set p low enough to induce full cooperation in the RR setting. This works fine in dichotomous systems (e.g. “sweet spot”). 2) Or we can include estimates for Ta <1 and Tb <1 in either the DR model ( T ) and the RR model ( T ’ or “T prime”). In higher order systems this sort of simulation becomes more important. 3) Regarding assumptions for values of T or T ’ , what if we’re assessing ALCOHOL consumption magnitude or frequency? Might the direction of “stigmatizing” be less predictable and linked to social groupings? Might *both* the no-alcohol and “excessive alcohol” groups carry some stigma? Member of (stigmatizing) A? yes yes π Ta (1- π )(1- Tb ) a lie by B no no π (1- Ta ) (1- π )( Tb ) a lie by A Truth by A&B BIAS = π [ 1 - 1 -2]+[1- 1 ] = 0 , if truth VAR = [ π *1+(1- π )(1-1)][(1- π *1)-(1- π )(1-1)] = π *(1- π )/n under the assumption of truth (T=1), reduces to more familiar variance for X~Binomial( π ) as proportion E[ π ^] = π * 1 +[(1- π )(1- 1 ) = π , if truth Bias = E( π ^- π )=E( π ^) -E( π ) = E( π ^) - π = π (Ta)+1- π -Tb+ π (Tb)- π](https://image.slidesharecdn.com/rrtrh51-12706549963629-phpapp02/85/Multinomial-Model-Simulations-11-320.jpg)

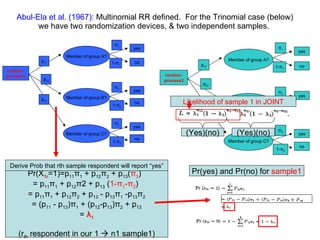
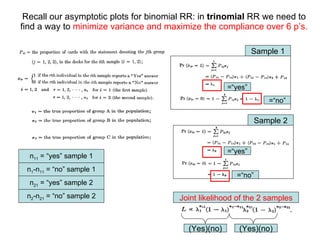








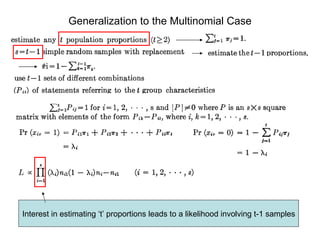
![For the multinomial generalization one gets a s ystem of equations that show expectation and variance of the [ s x 1 ] vector of proportions ( π i )](https://image.slidesharecdn.com/rrtrh51-12706549963629-phpapp02/85/Multinomial-Model-Simulations-32-320.jpg)