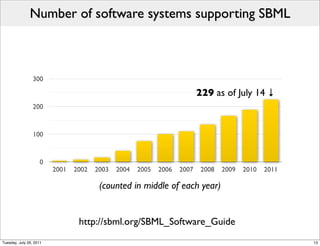
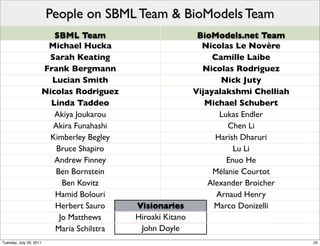
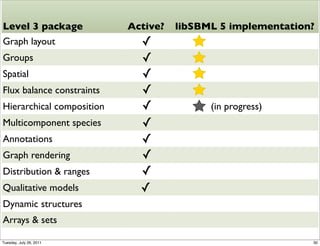
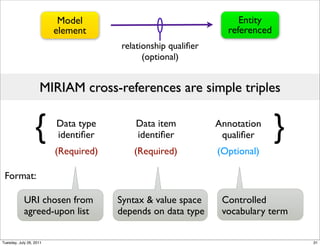
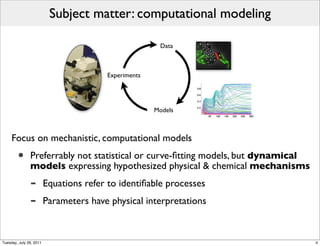
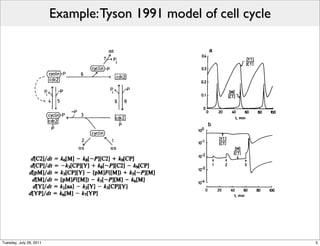
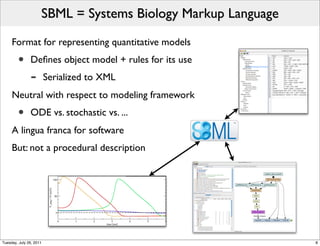
The document discusses the Systems Biology Markup Language (SBML), a format for representing quantitative models in computational biology, emphasizing the need for effective sharing of models and reproducibility in simulations. It outlines the evolution of SBML, its specifications, and the community's ongoing efforts to develop interoperable standards and software tools. Key lessons learned from the SBML development process highlight the importance of stakeholder engagement, simplicity, and formal procedures.




![Basic SBML concepts are simple
The reaction is central: a process occurring at a given rate
• Participants are pools of entities (species)
f ([A],[B],[P ],...)
na A + nb B − − − − − − → np P
−−−−−−
f (...)
nc C −−
−→ nd D + ne E + nf F
.
.
.
Models can further include:
• Other constants & variables • Unit definitions
• Compartments • Annotations
• Explicit math
• Discontinuous events
Tuesday, July 26, 2011 9](https://image.slidesharecdn.com/mhucka-cabig-icr-2011-07-120706232709-phpapp02/85/SBML-the-Systems-Biology-Markup-Language-9-320.jpg)
![Basic SBML concepts are simple
The reaction is central: a process occurring at a given rate
• Participants are pools of entities (species) Can be anything
conceptually
f ([A],[B],[P ],...)
na A + nb B − − − − − − → np P
−−−−−− compatible
f (...)
nc C −−
−→ nd D + ne E + nf F
.
.
.
Models can further include:
• Other constants & variables • Unit definitions
• Compartments • Annotations
• Explicit math
• Discontinuous events
Tuesday, July 26, 2011 9](https://image.slidesharecdn.com/mhucka-cabig-icr-2011-07-120706232709-phpapp02/85/SBML-the-Systems-Biology-Markup-Language-10-320.jpg)