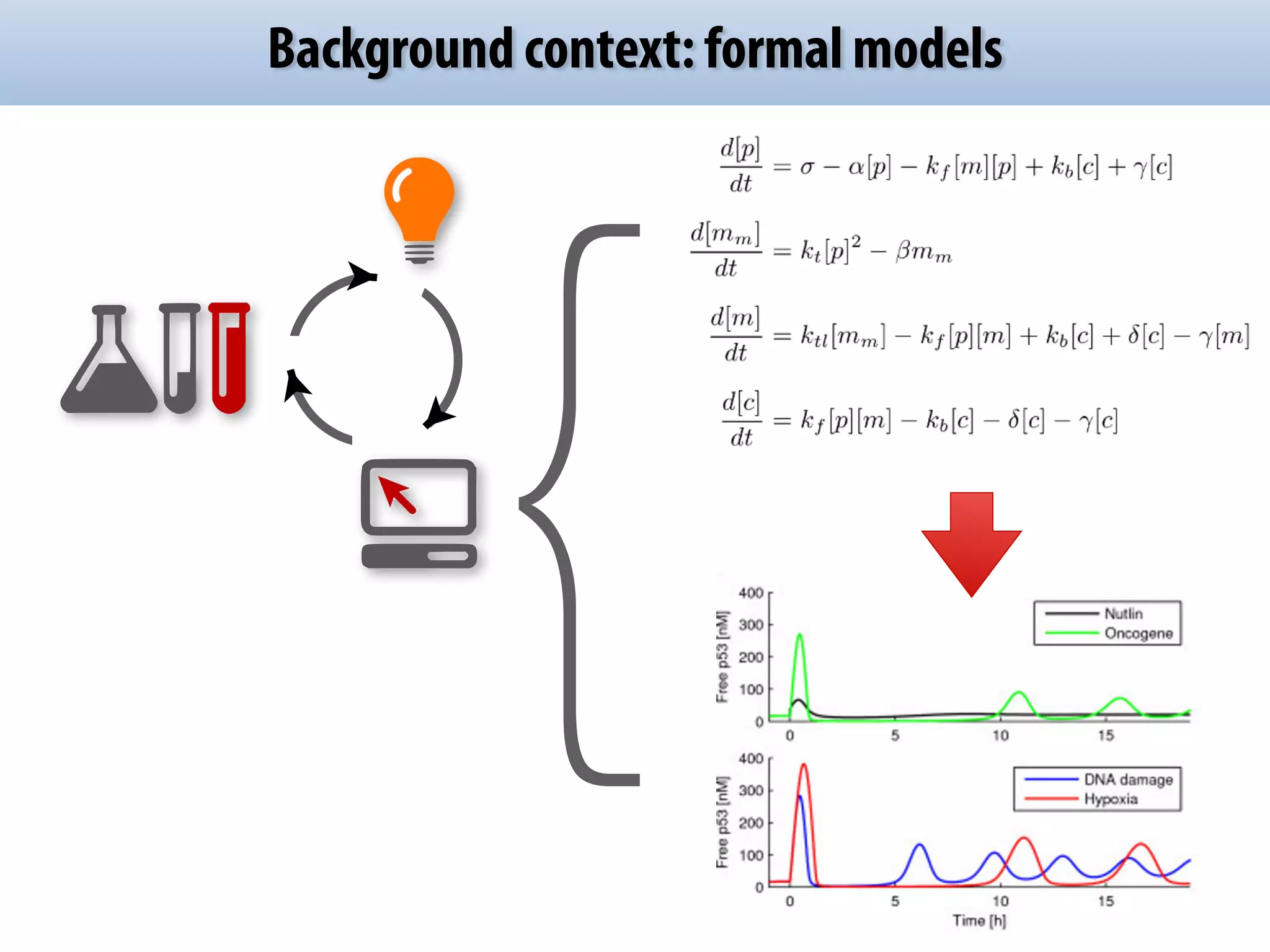
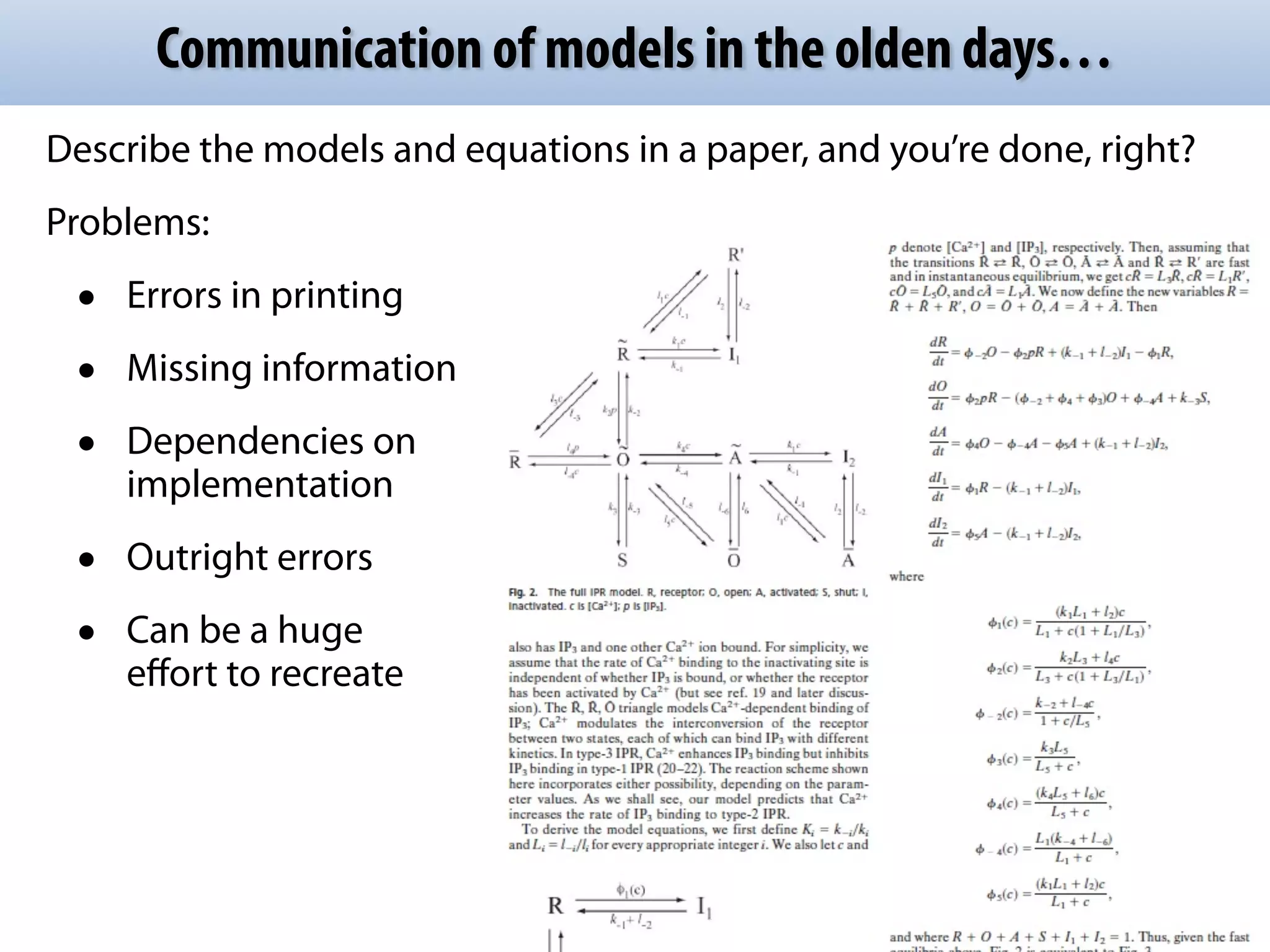
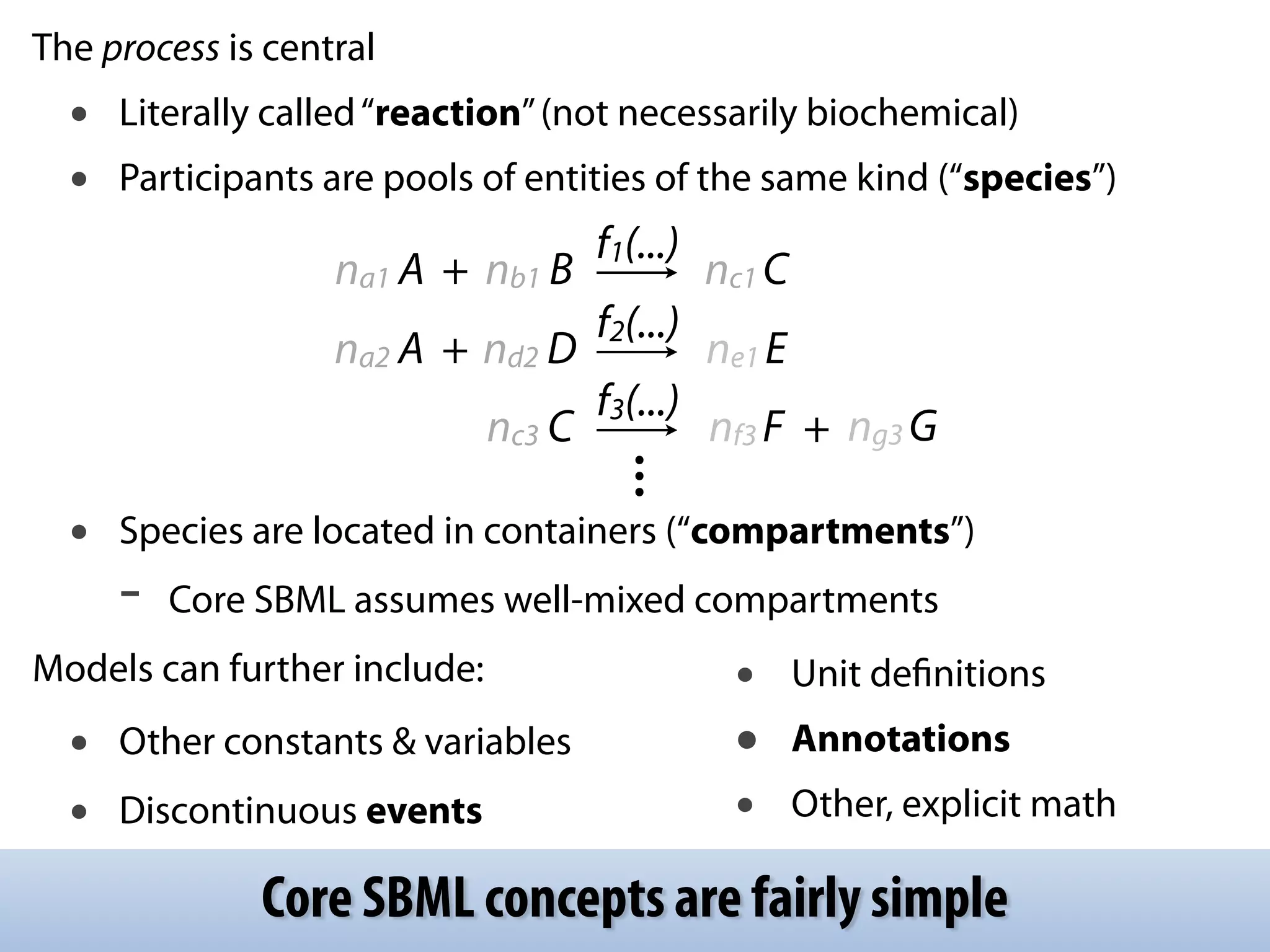
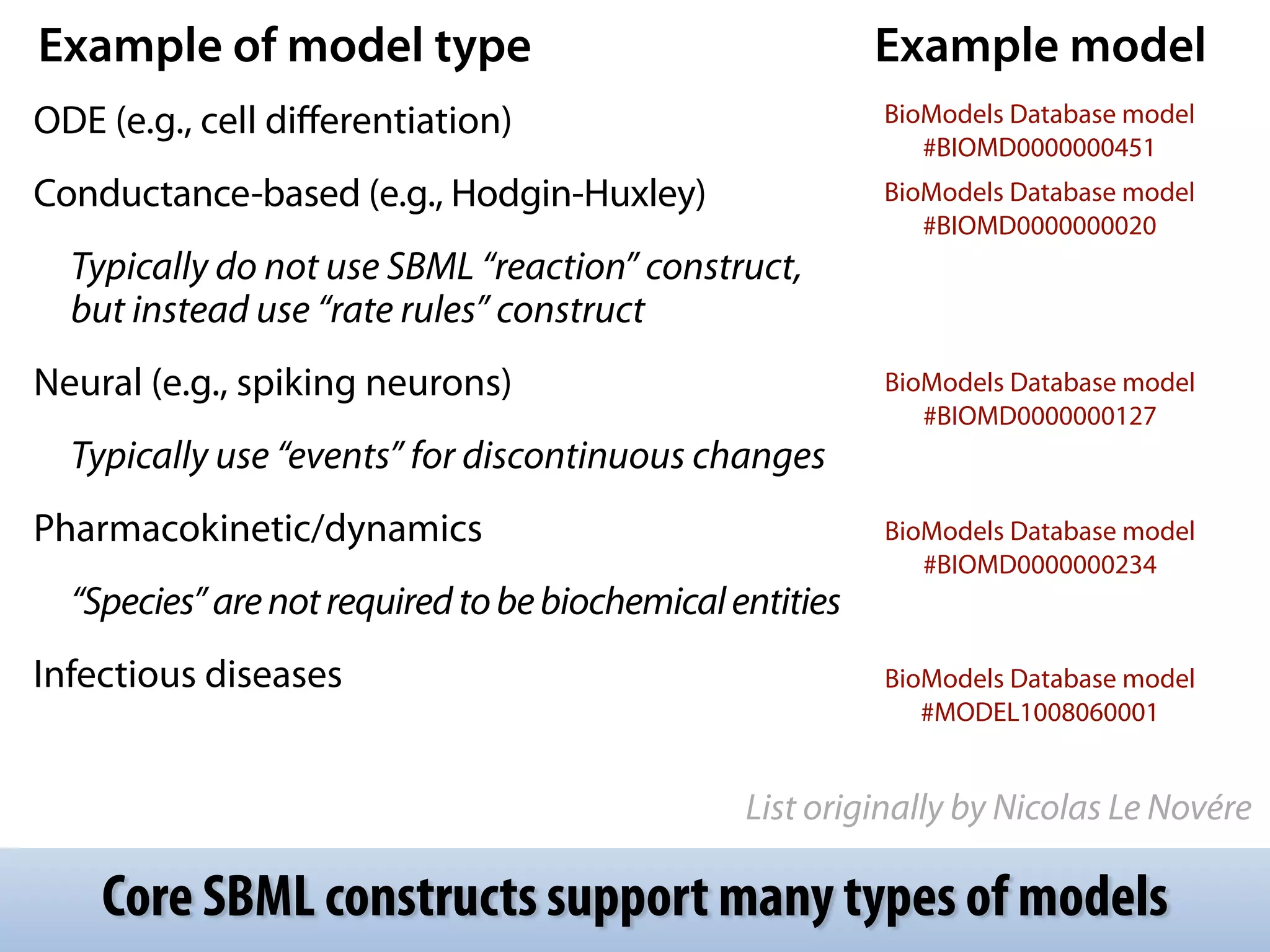
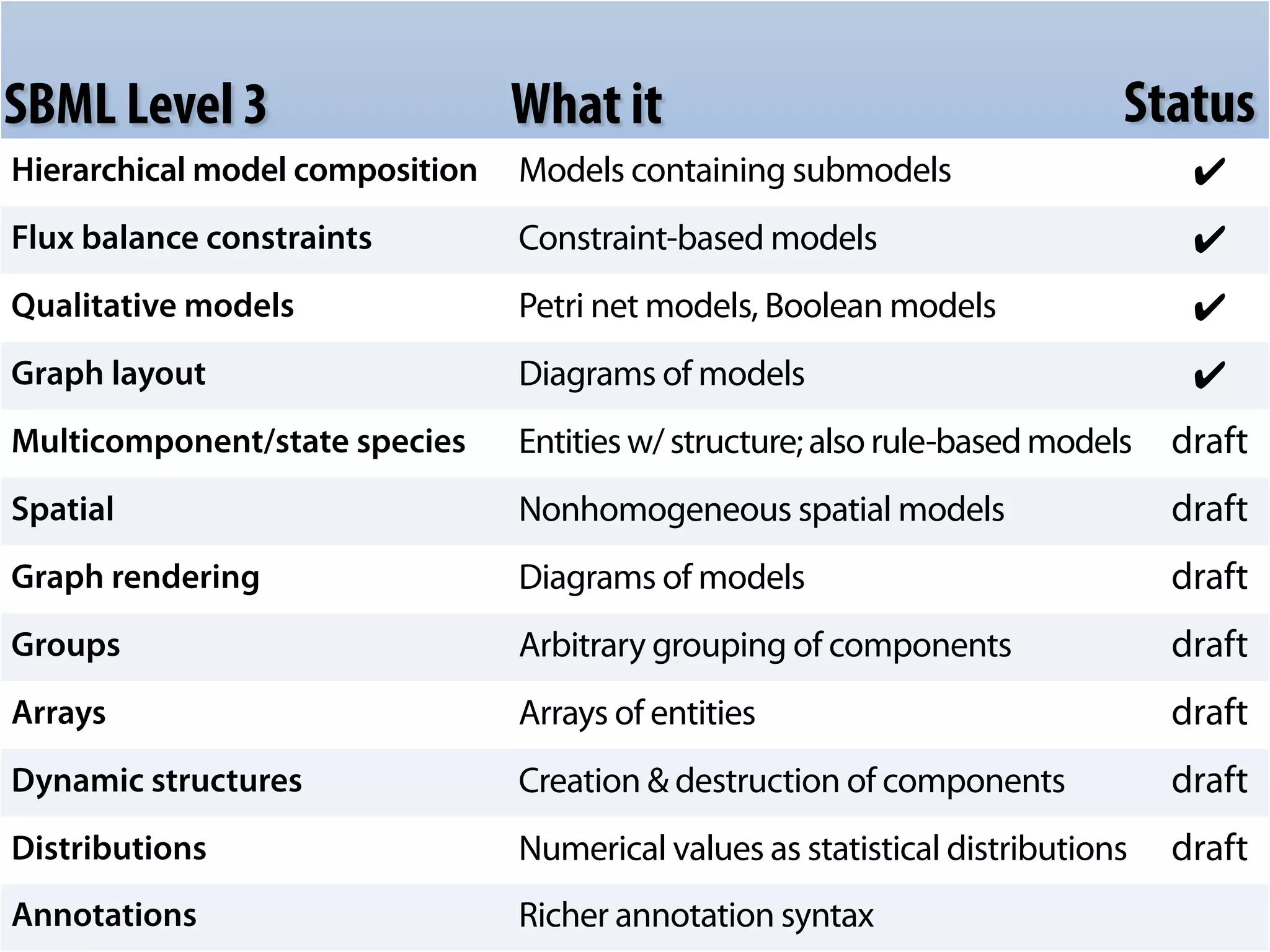
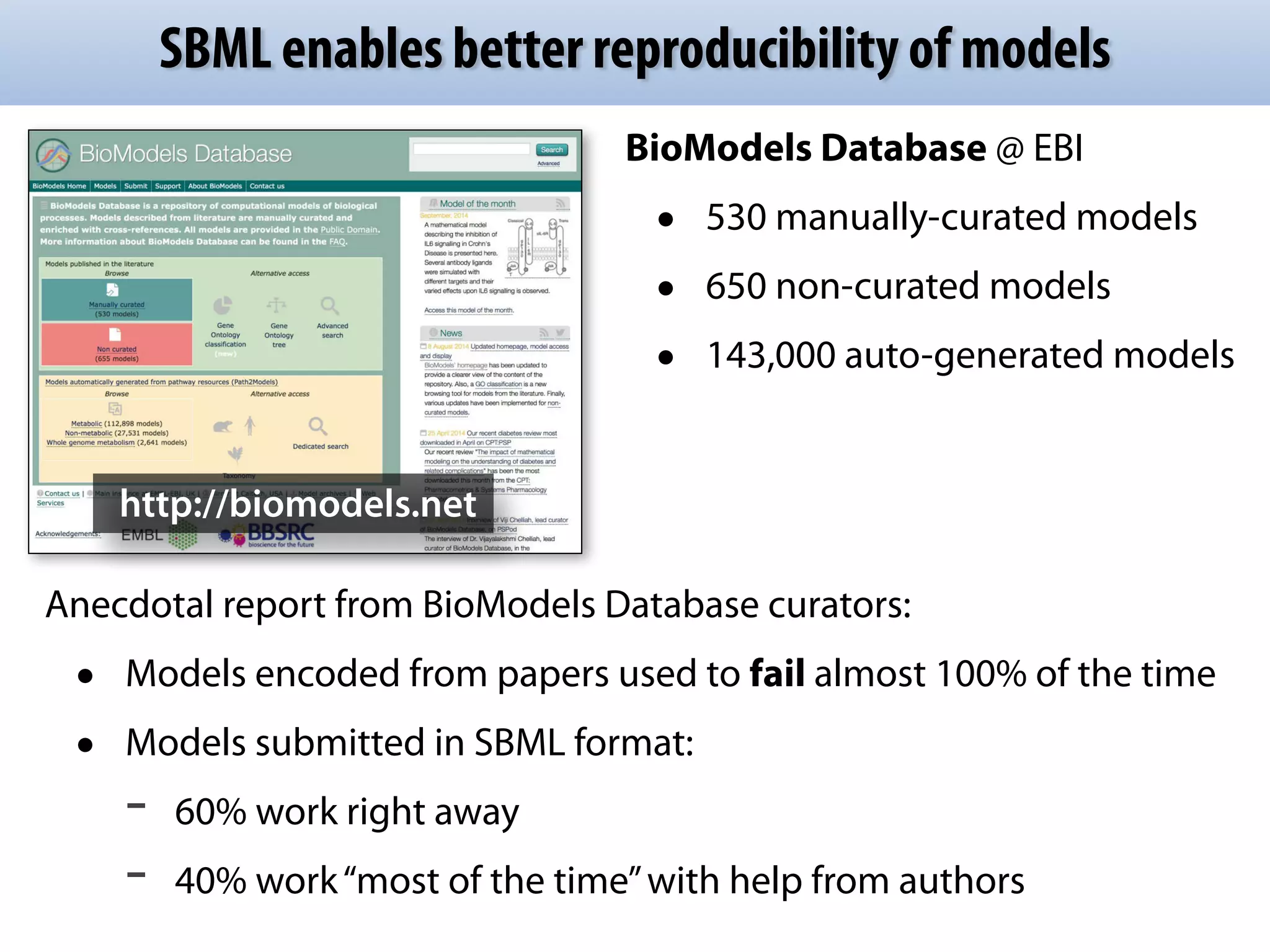
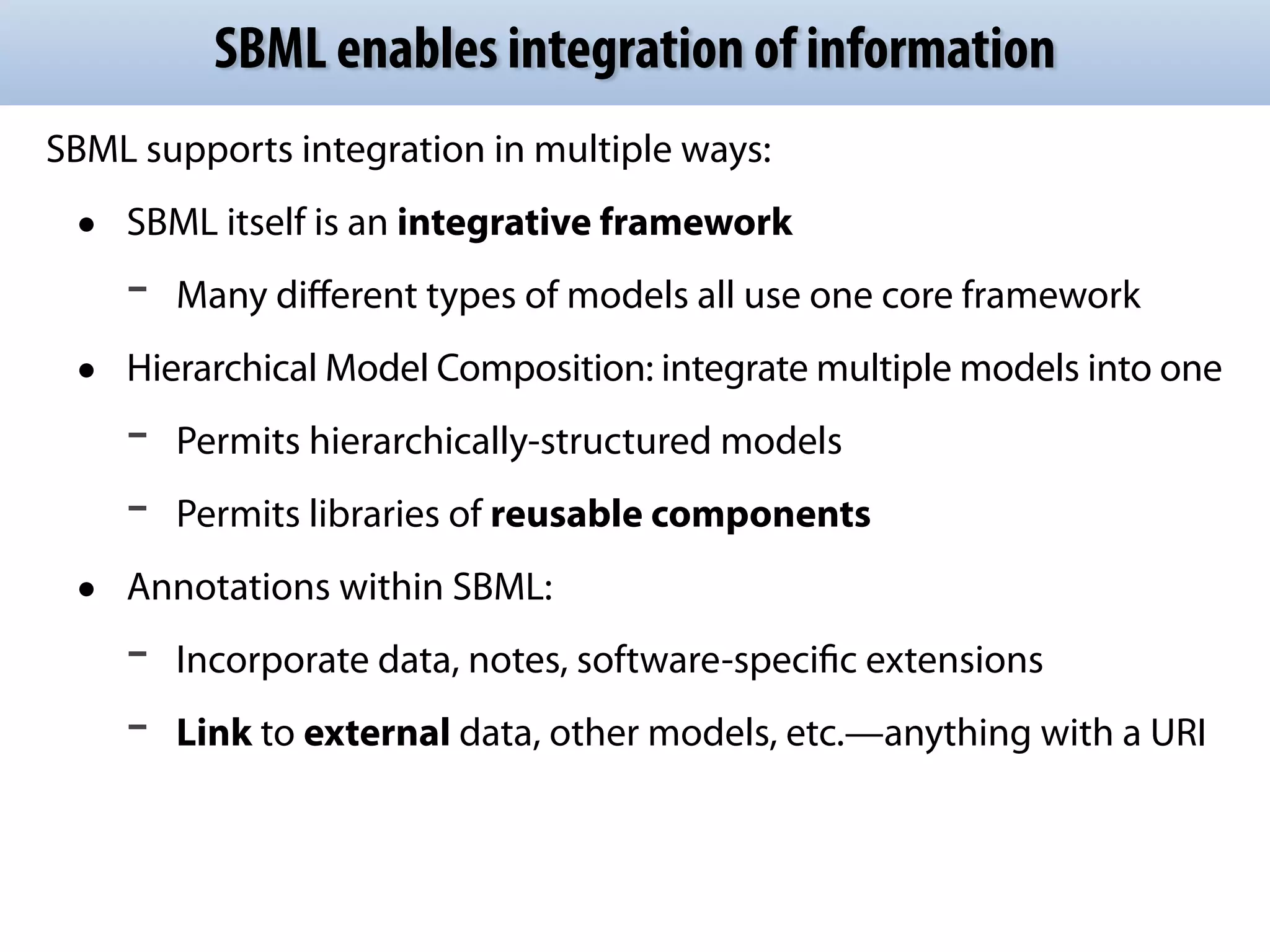
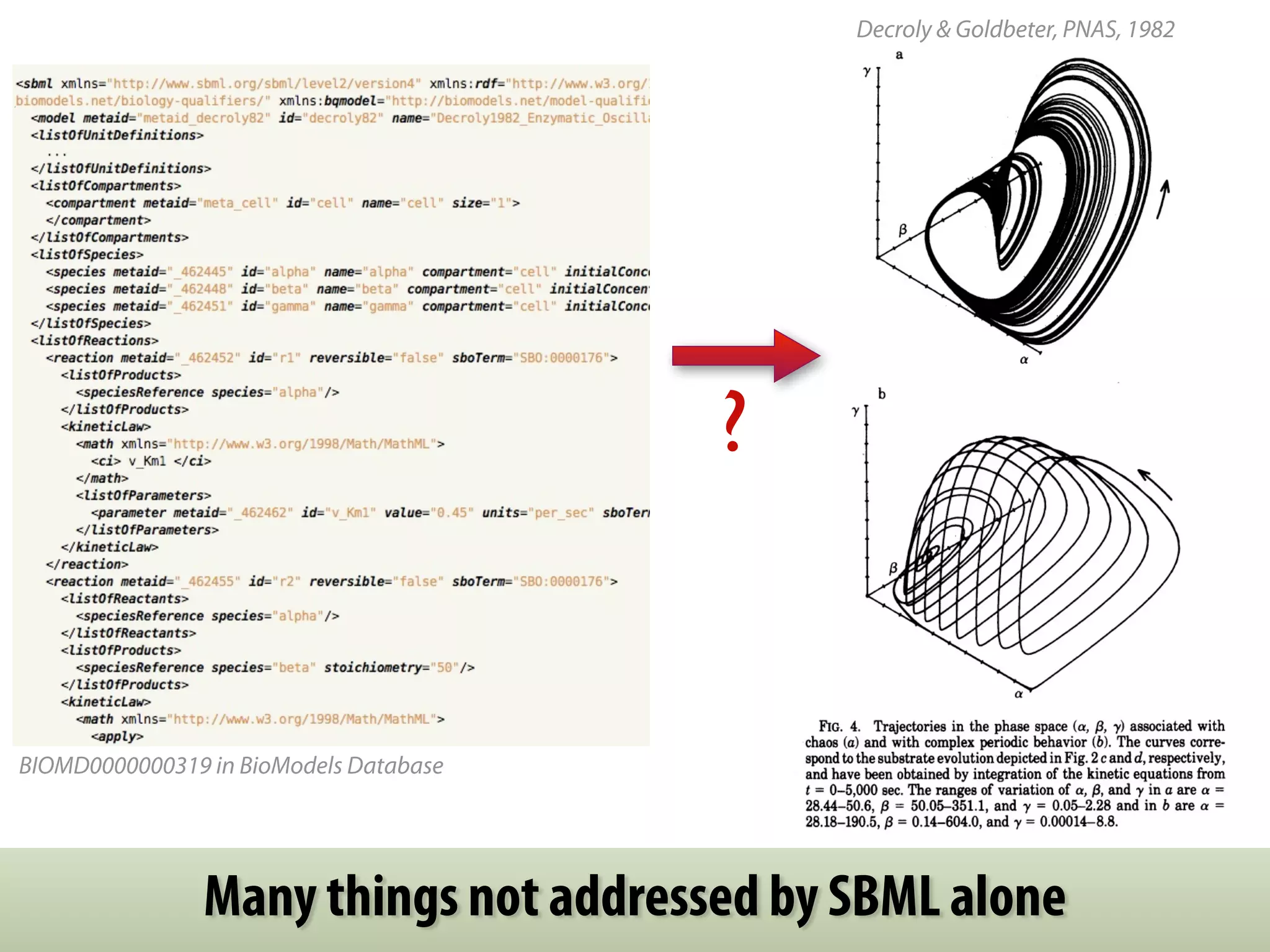
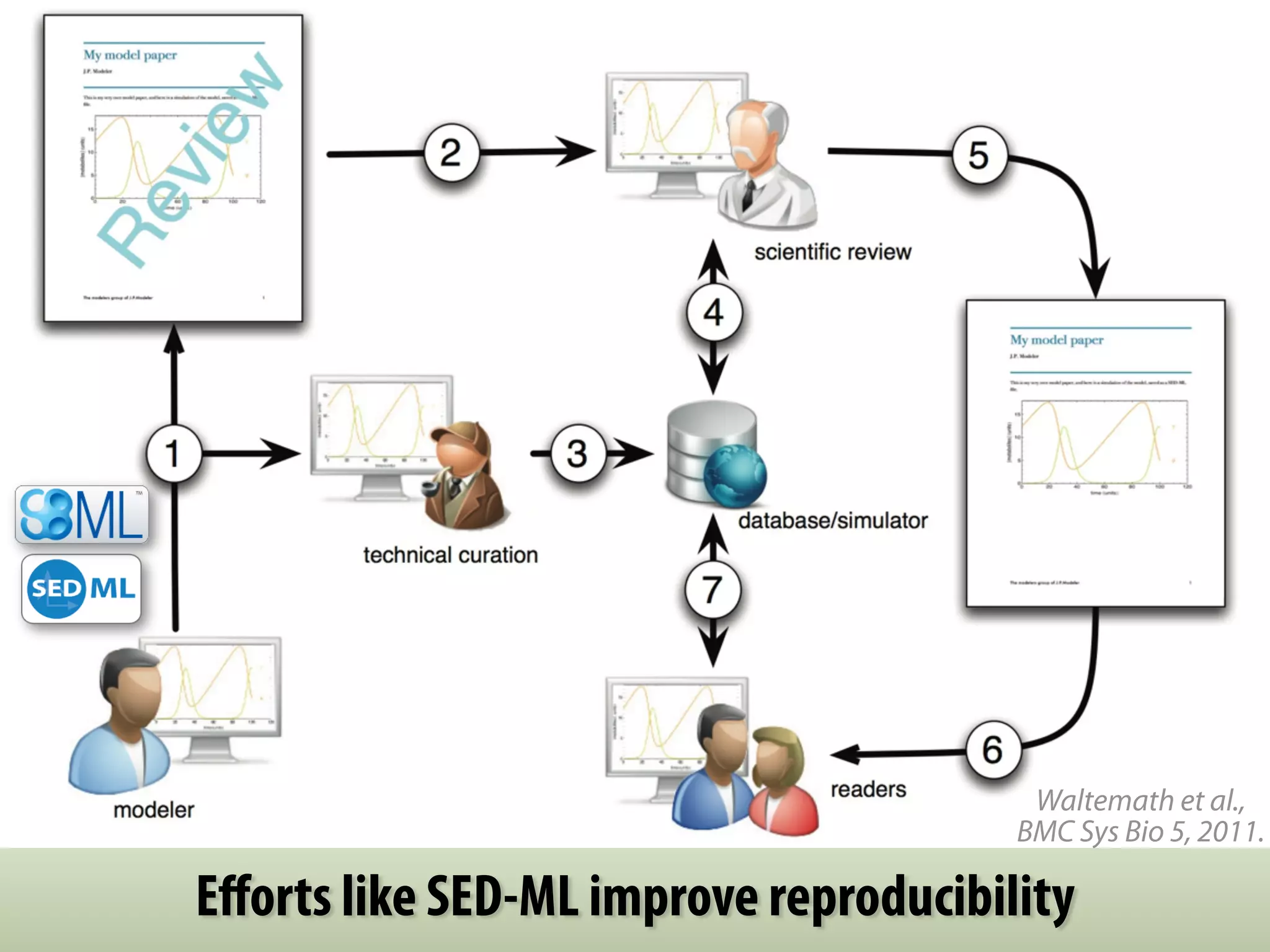
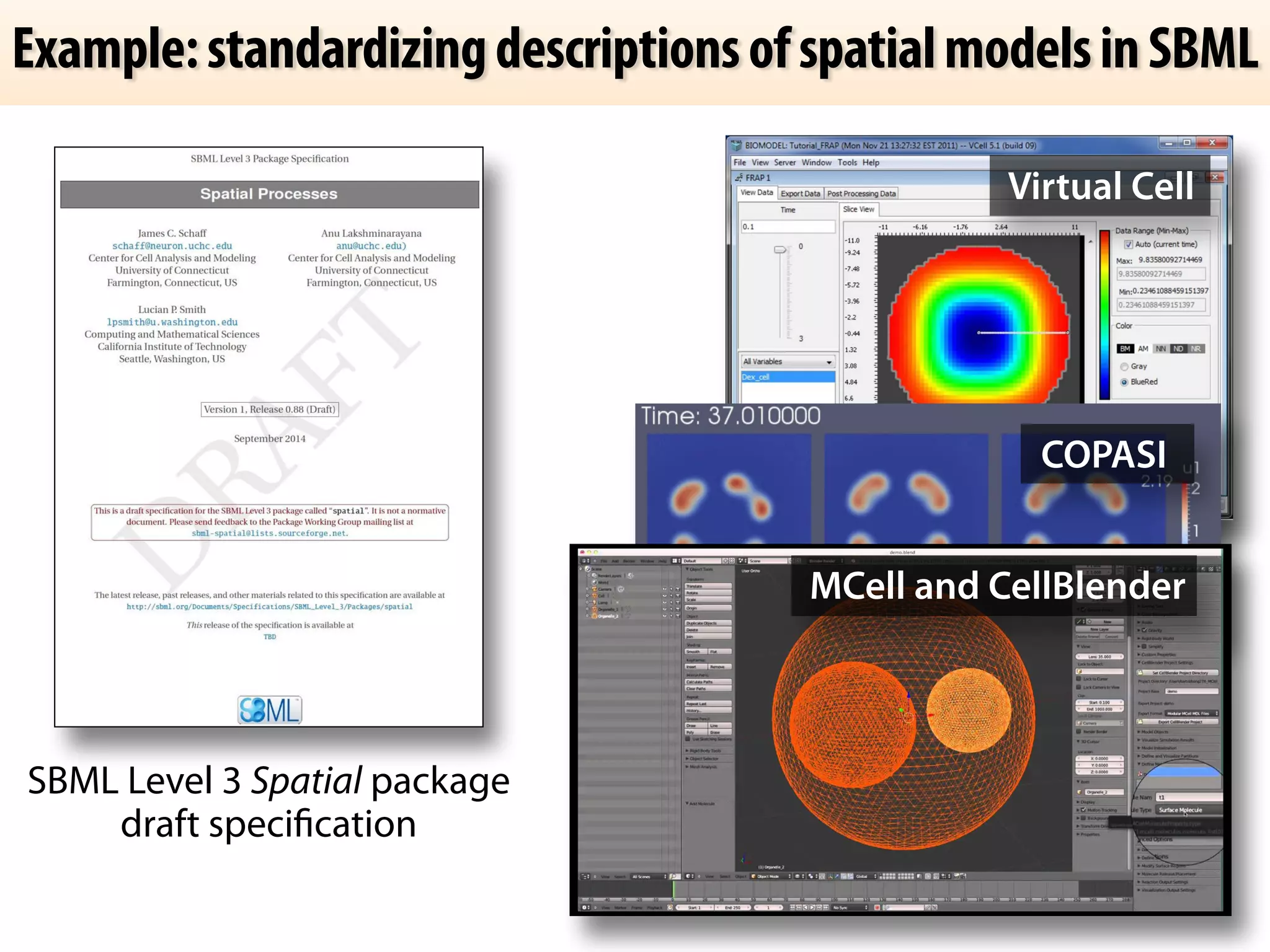
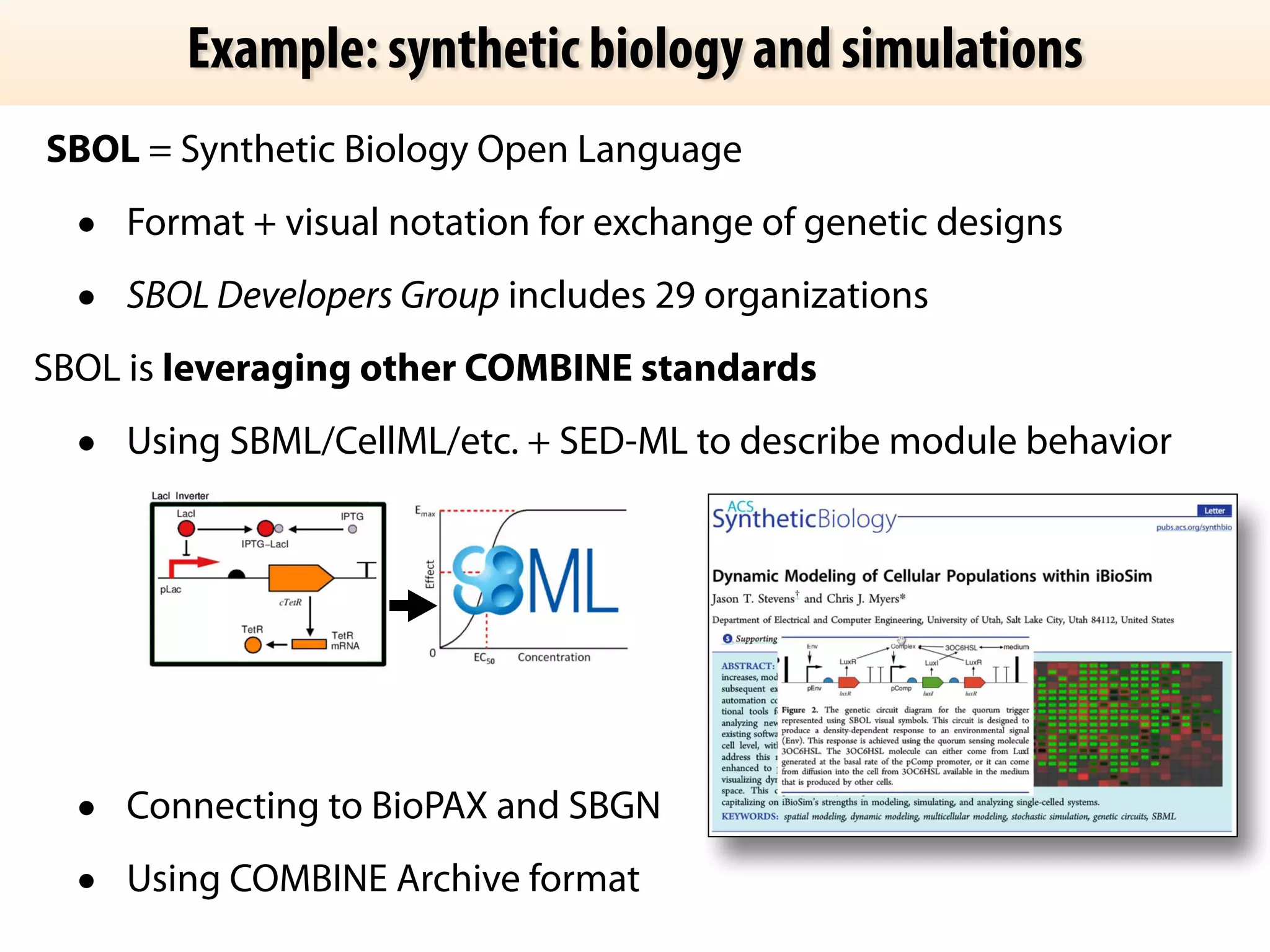
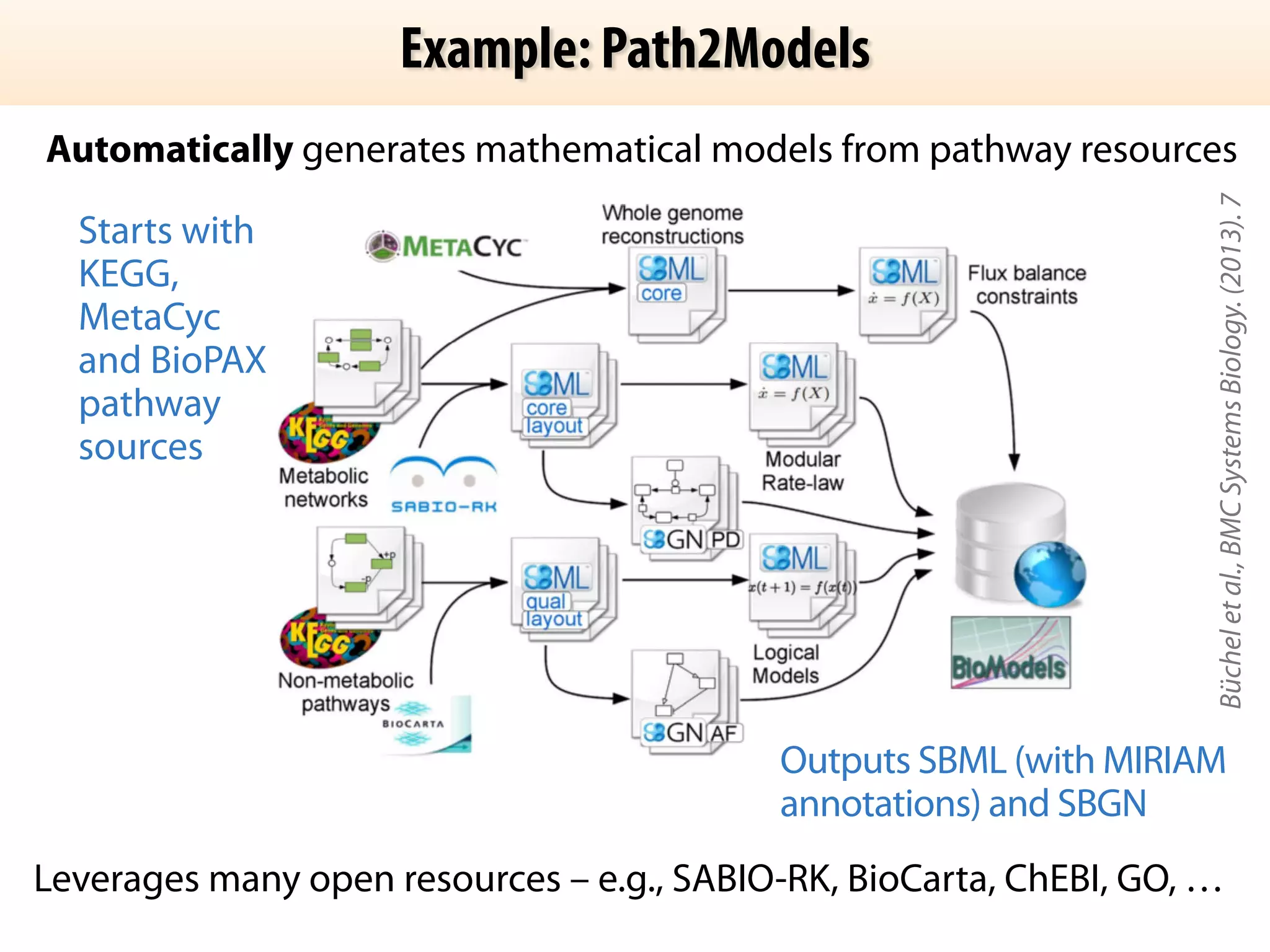
The document discusses the significance of the Systems Biology Markup Language (SBML) in enhancing the reproducibility of computational research in systems biology. It highlights the challenges faced in model representation and the integration of various software tools, emphasizing the ongoing standardization efforts through initiatives like the COMBINE network. Additionally, it outlines a range of available software for modeling and simulation while noting the extensive resources, including hundreds of standardized models and their applications.