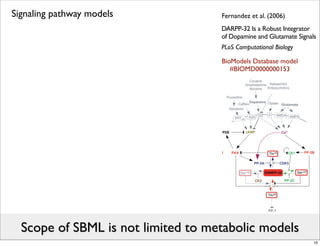
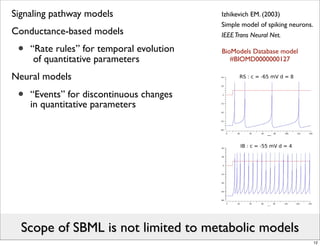
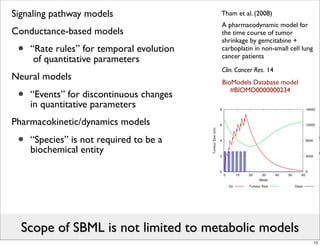
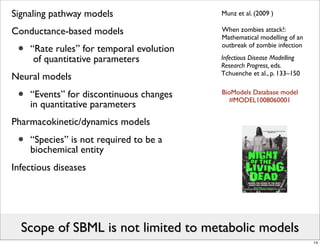
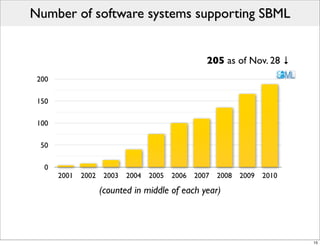
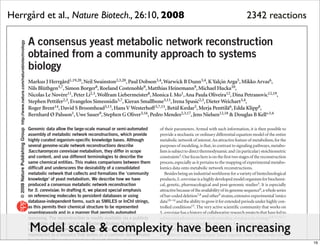
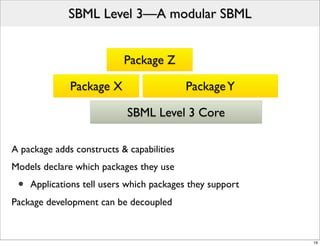
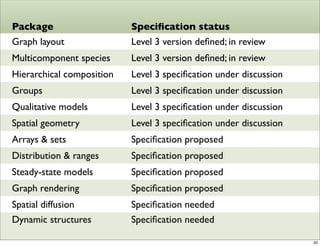
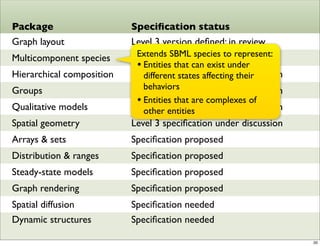
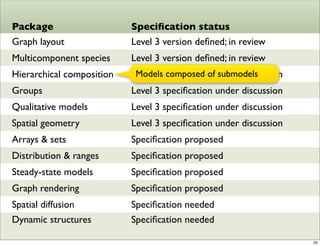
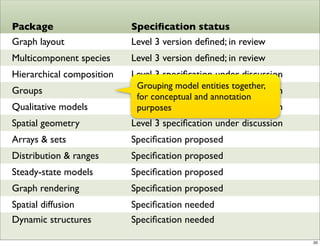
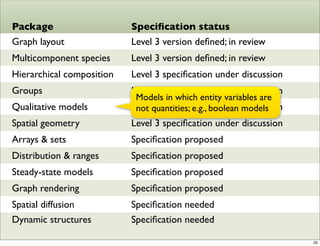
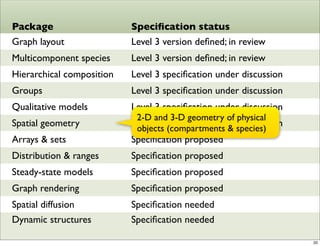
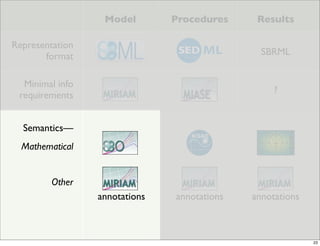
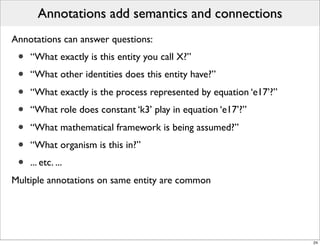
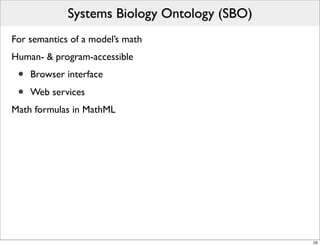
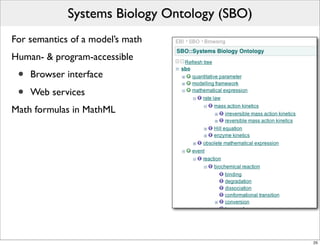
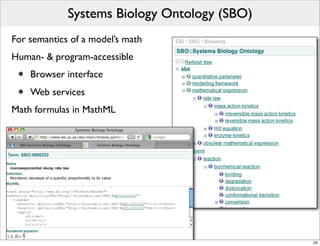
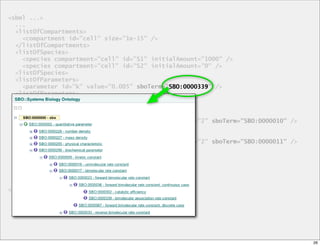
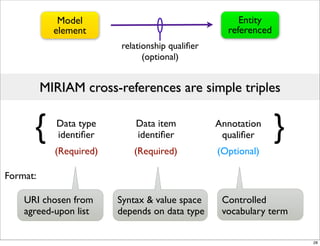
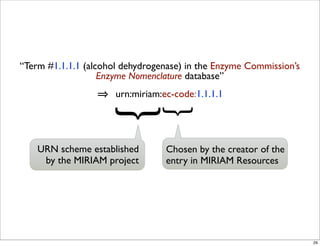
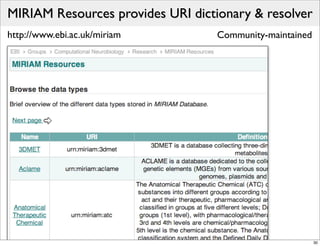
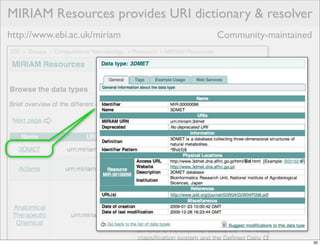
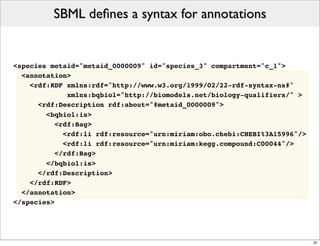
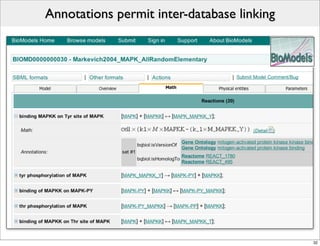
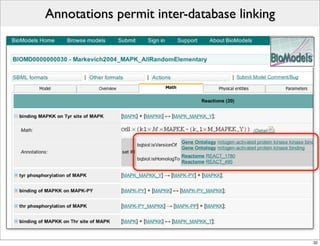
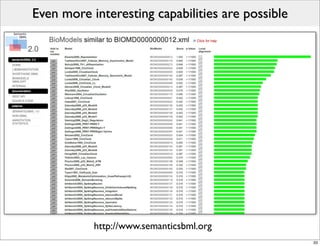
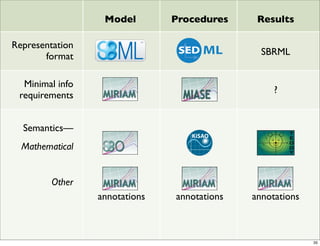
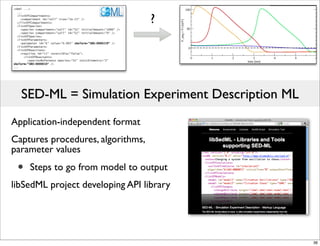
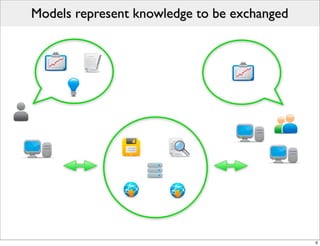
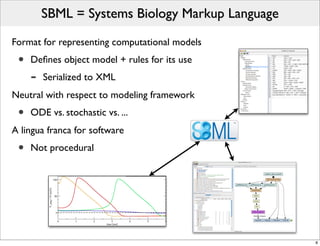
The document discusses Systems Biology Markup Language (SBML), a format for representing computational models of biological processes. SBML allows models to be exchanged between different software applications and defines concepts like species, compartments, reactions, and parameters. It aims to serve as a common language for software in systems biology. The document outlines some basic SBML concepts and notes that the scope of SBML is not limited to metabolic models, but can also represent signaling pathways, neural models, pharmacokinetic models, and other types of models. It discusses how SBML continues to evolve through new Levels and Packages to support additional model constructs and capabilities.




![Basic SBML concepts are simple
The reaction is central: a process occurring at a given rate
• Participants are pools of entities (species)
f ([A],[B],[P ],...)
na A + nb B − − − − − − → np P
−−−−−−
f (...)
nc C −−
−→ nd D + ne E + nf F
.
.
.
Models can further include:
• Other constants & variables • Unit definitions
• Compartments • Annotations
• Explicit math
• Discontinuous events
9](https://image.slidesharecdn.com/mhucka-cosbi-2010-12-120706233008-phpapp01/85/Finding-common-ground-between-modelers-and-simulation-software-in-systems-biology-8-320.jpg)
![Basic SBML concepts are simple
The reaction is central: a process occurring at a given rate
• Participants are pools of entities (species) Can be anything
conceptually
f ([A],[B],[P ],...)
na A + nb B − − − − − − → np P
−−−−−− compatible
f (...)
nc C −−
−→ nd D + ne E + nf F
.
.
.
Models can further include:
• Other constants & variables • Unit definitions
• Compartments • Annotations
• Explicit math
• Discontinuous events
9](https://image.slidesharecdn.com/mhucka-cosbi-2010-12-120706233008-phpapp01/85/Finding-common-ground-between-modelers-and-simulation-software-in-systems-biology-9-320.jpg)