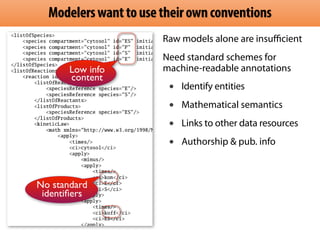
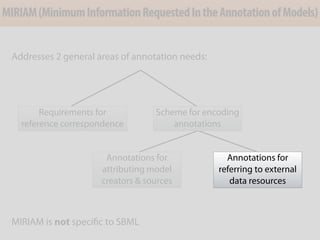
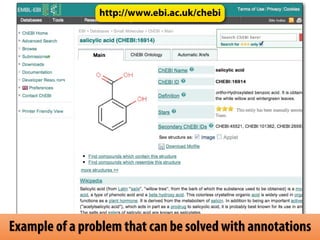
The document discusses the creation and importance of the Systems Biology Markup Language (SBML) in facilitating the communication and reproducibility of computational models in biological research. It highlights shared efforts with MIRIAM and SED-ML to improve model annotations and integrate systems biology standards through community collaboration. Additionally, it outlines the evolution and future development of SBML while reflecting on lessons learned from past standardization efforts.