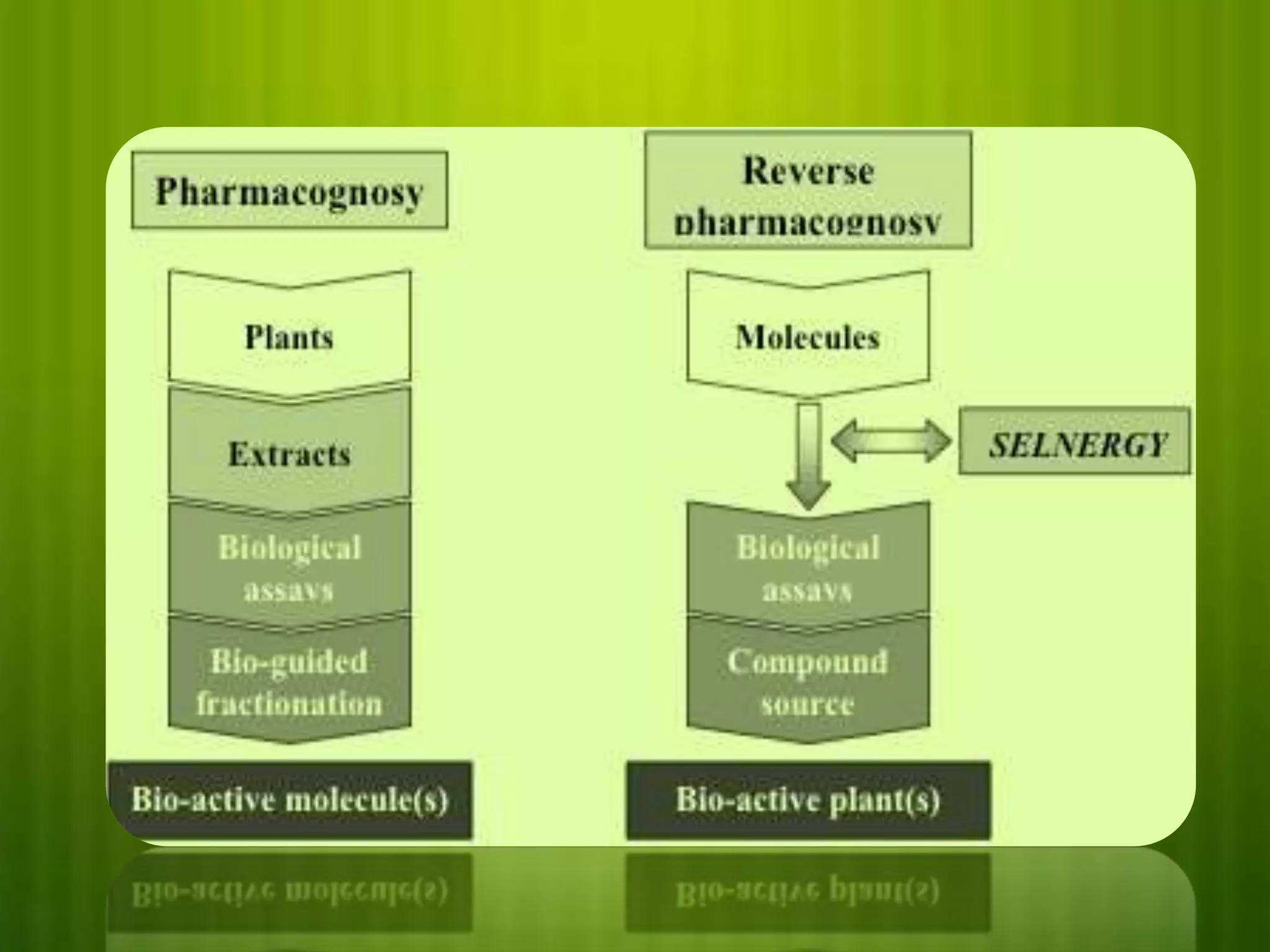
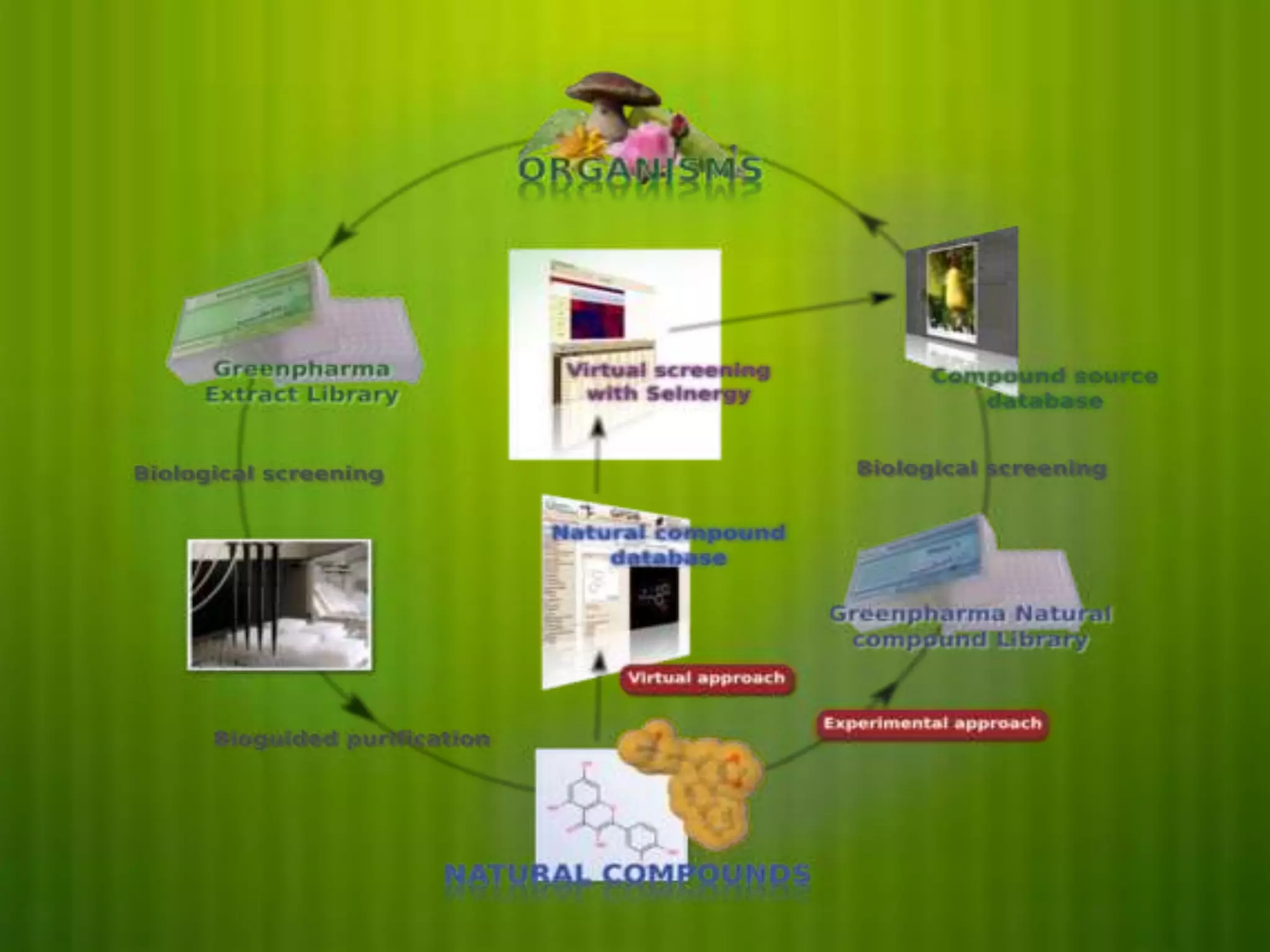
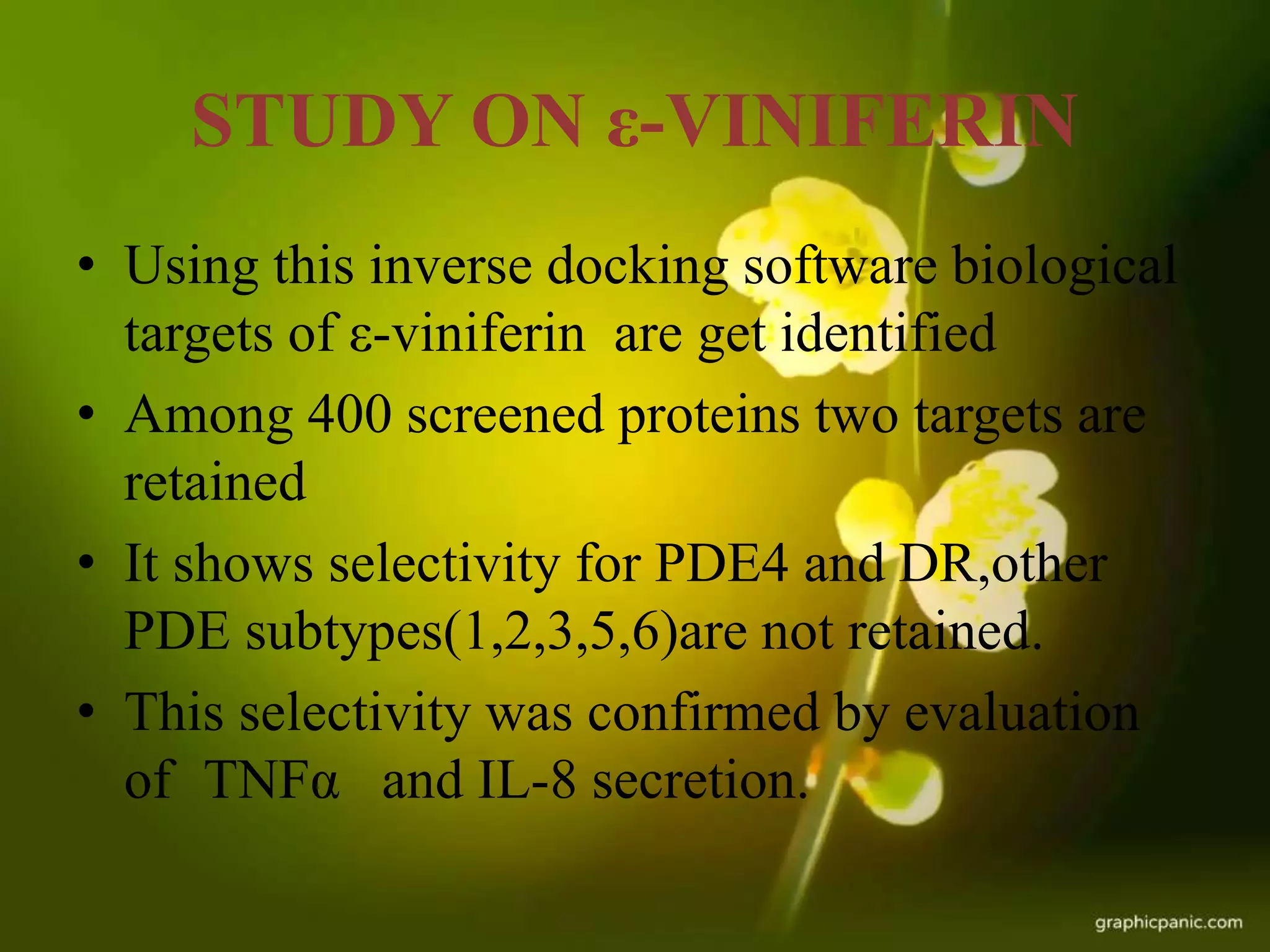
This document discusses reverse pharmacognosy, an approach that starts with a biologically active molecule and uses computational tools to identify potential plant sources of that molecule, rather than the conventional approach of screening many plant extracts. It describes Selenergy software that predicts molecule-protein interactions and identifies biological targets. Case studies are presented on identifying targets of ε-viniferin from grapes, meranzin from Limnocitrus, and tofisopam. Reverse pharmacognosy can accelerate drug discovery, allow drug repositioning, and decrease plant usage compared to conventional screening methods.