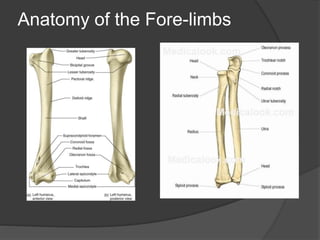
This document discusses regeneration in different animal groups. It explains that regeneration involves reactivating development to restore missing tissue. Regeneration occurs through stem cell mediated regeneration, epimorphosis, morphallaxis, and compensatory regeneration. Epimorphosis involves dedifferentiation and redifferentiation, as seen in salamander limb regeneration. Compensatory regeneration uses differentiated cells and is seen in liver and zebrafish heart regeneration. Regeneration also relies on signaling gradients that establish polarity and pattern formation.