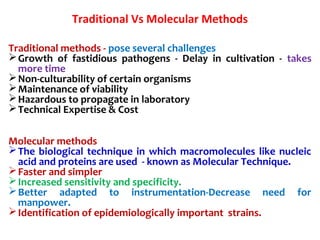
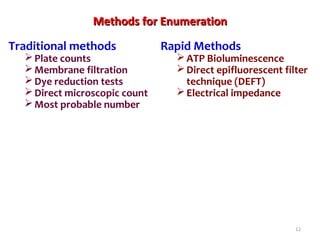
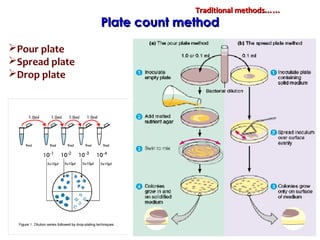
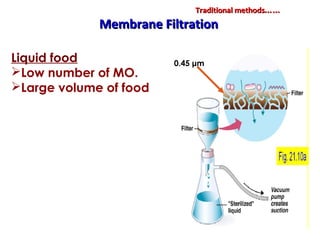
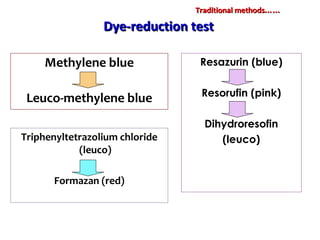
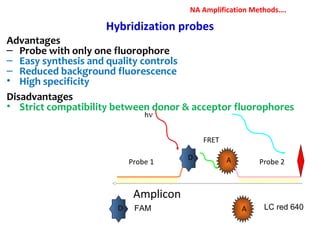
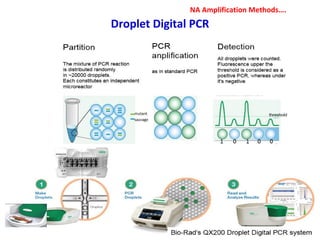
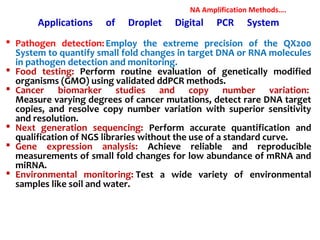
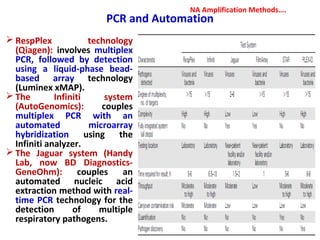
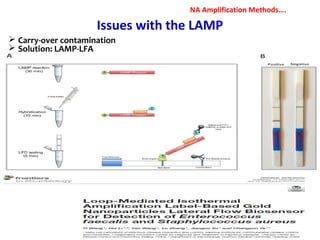
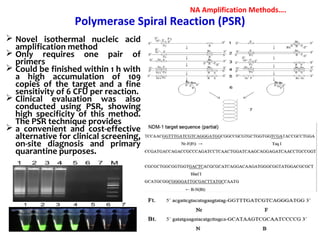
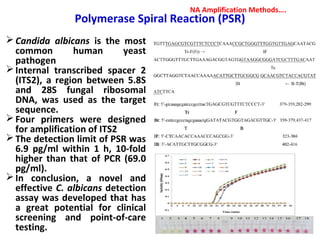
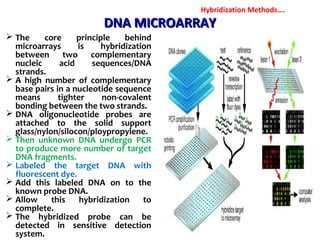
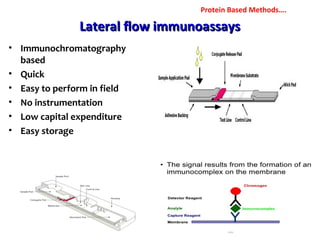
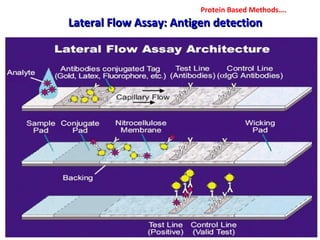
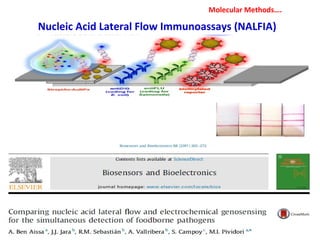
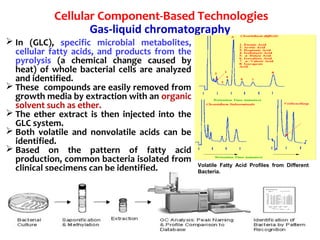
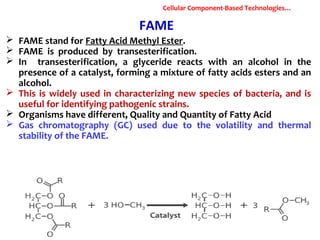
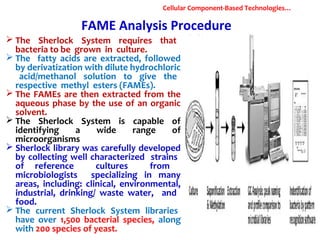
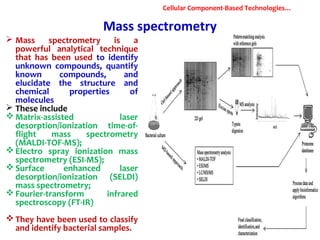
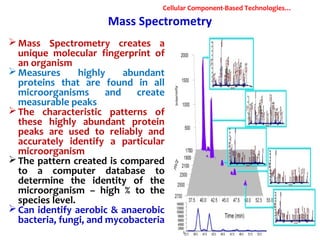
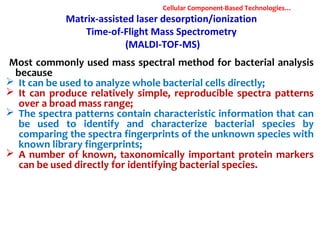
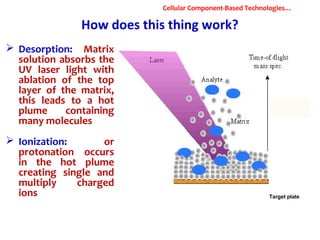
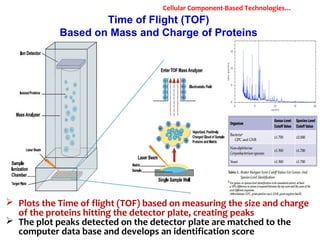
1. Molecular techniques provide faster and more accurate summaries of meat and meat products compared to traditional methods. They allow for identification of pathogens and toxins with increased sensitivity and specificity.
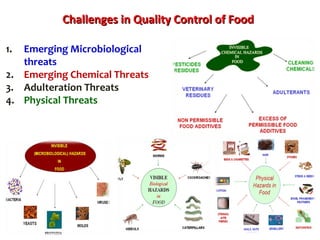
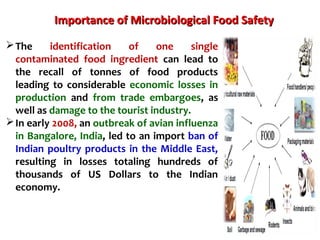
2. India has a large livestock population that contributes significantly to its agricultural GDP. However, ensuring food safety is challenging due to emerging microbiological and chemical threats.
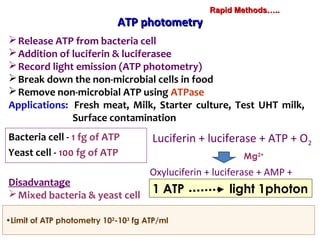
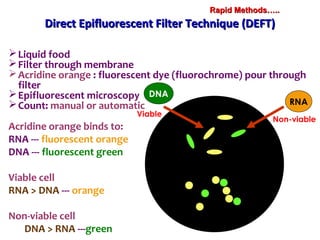
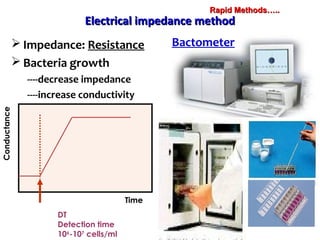
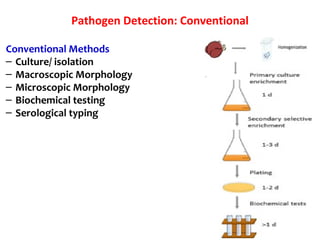
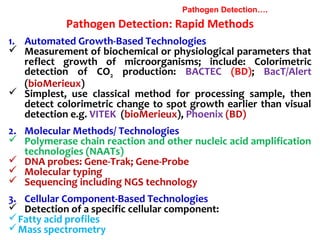
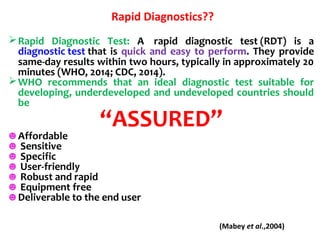
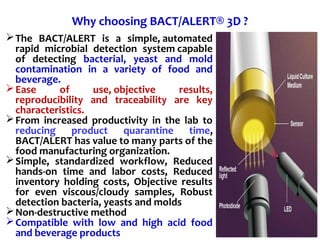
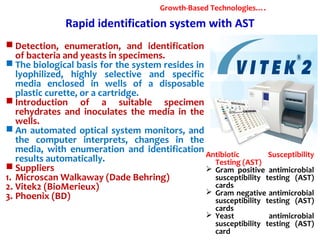
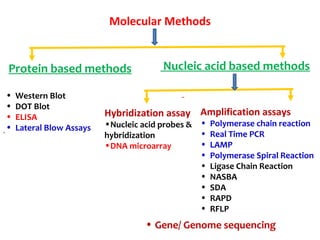
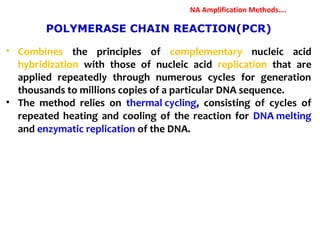
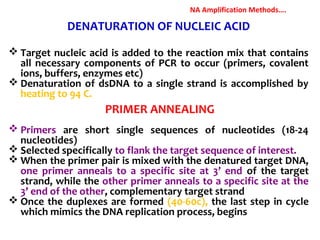
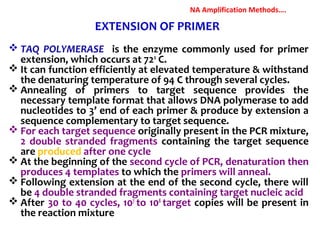
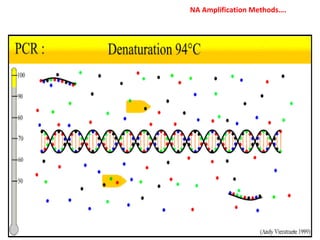
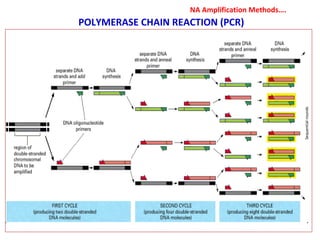
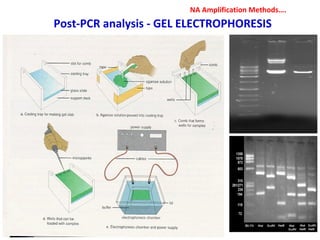
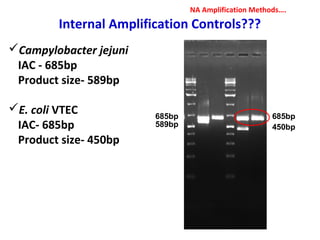
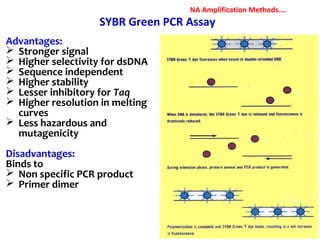
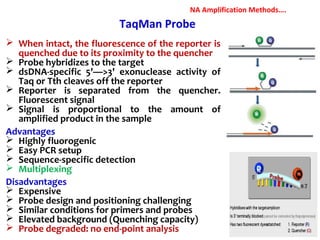
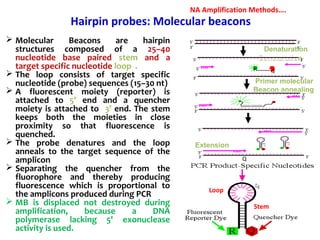
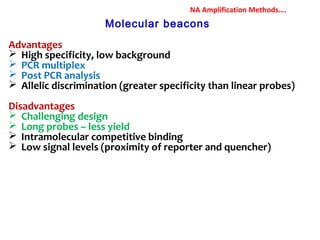
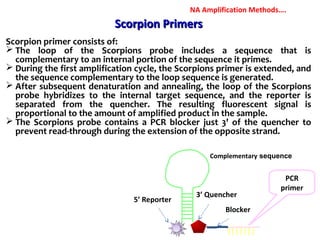
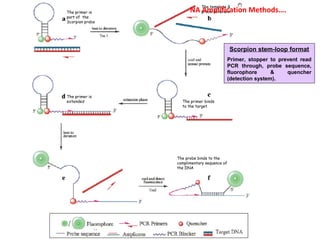
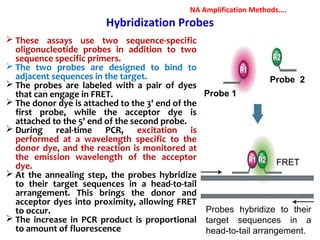
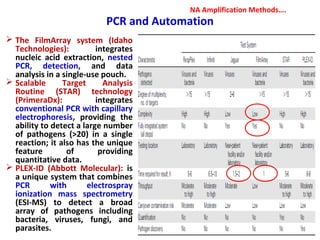
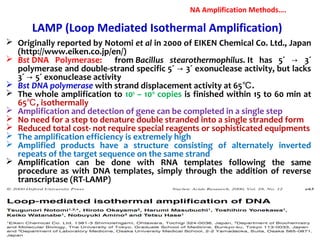
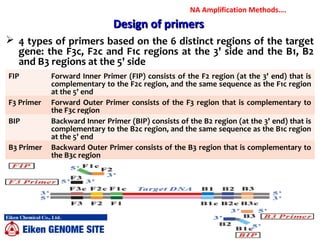
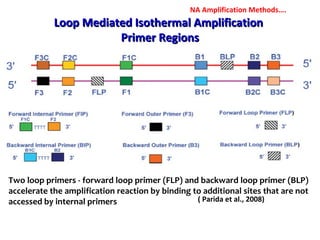
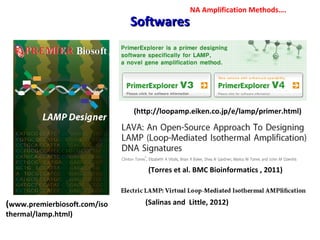
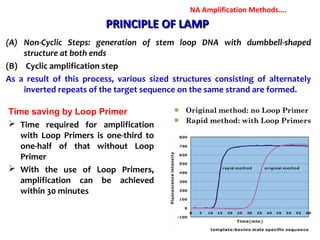
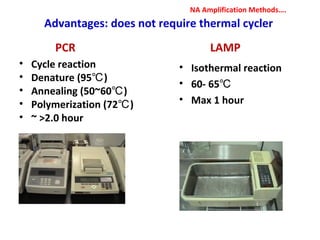
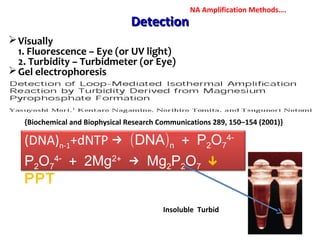
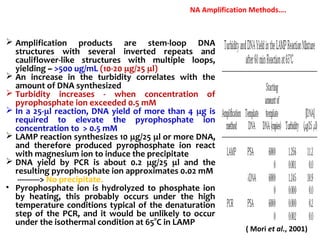
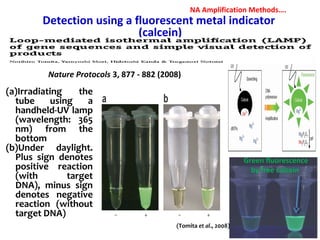
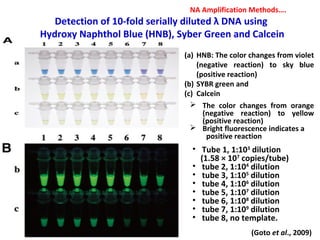
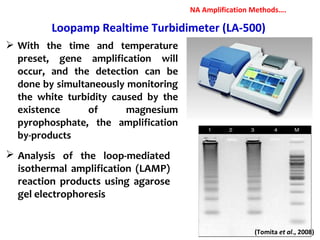
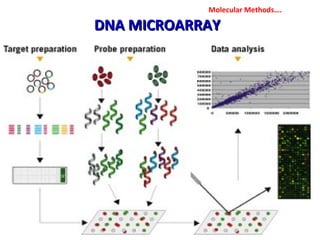
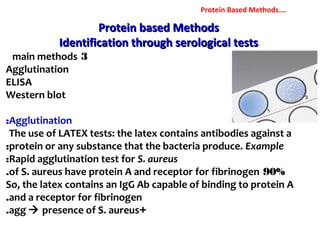
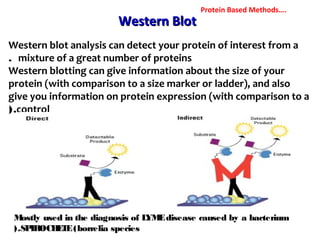
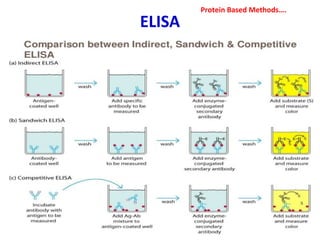
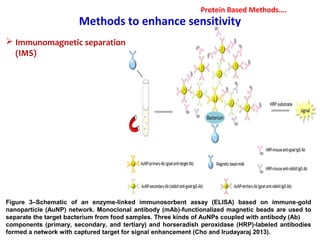
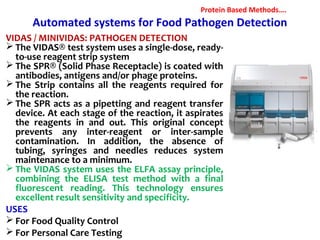
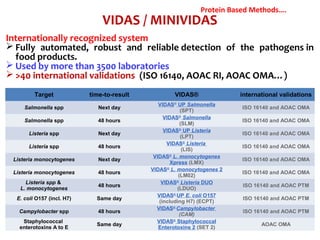
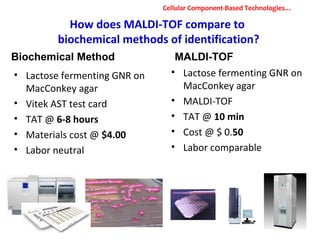
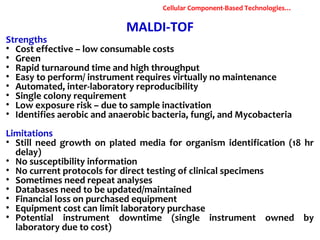
3. Rapid molecular detection methods like PCR and DNA probes are now used alongside automated growth-based techniques to quickly detect foodborne pathogens in meat. This allows for timely recalls and prevents economic losses from trade issues.