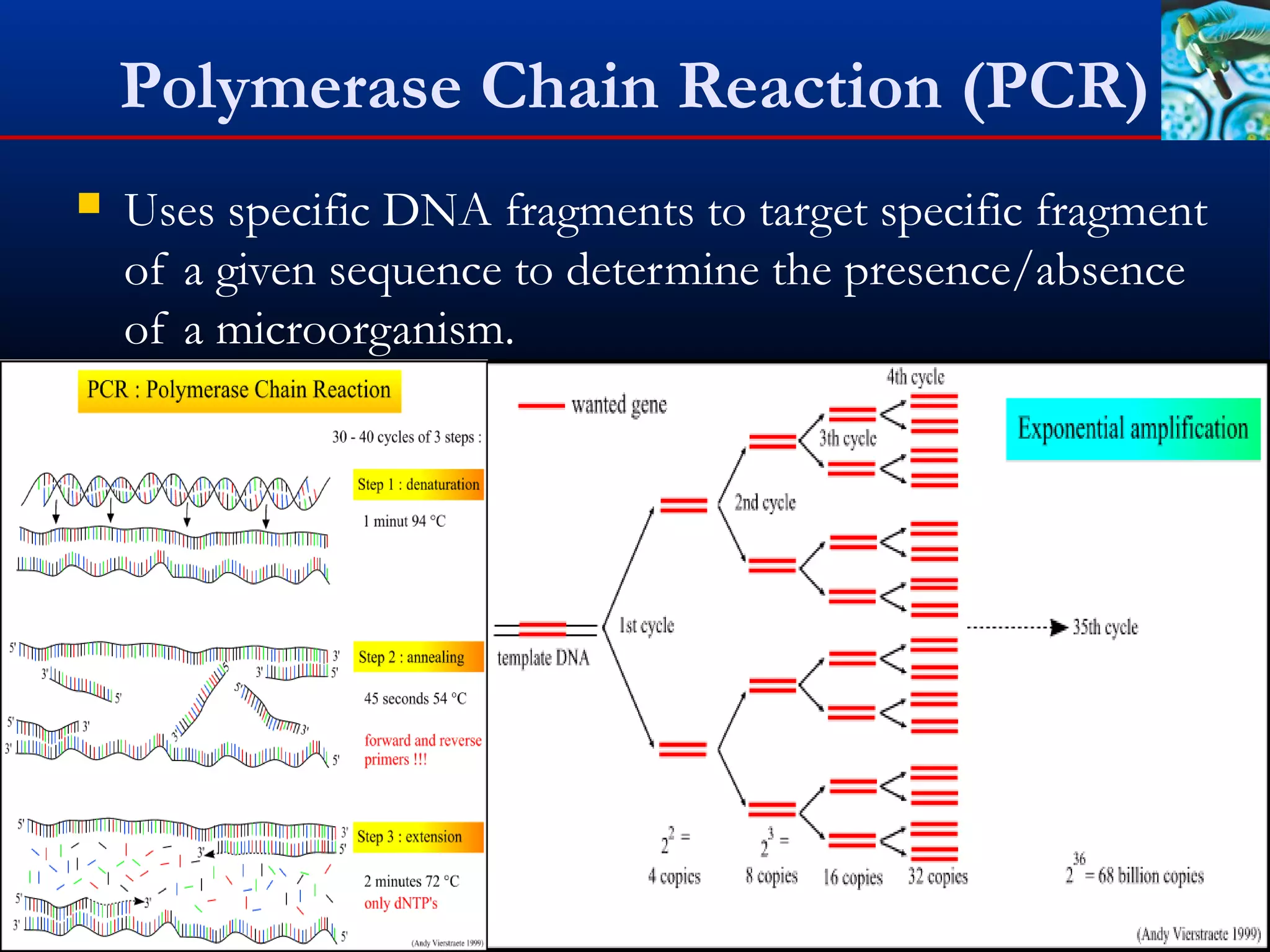
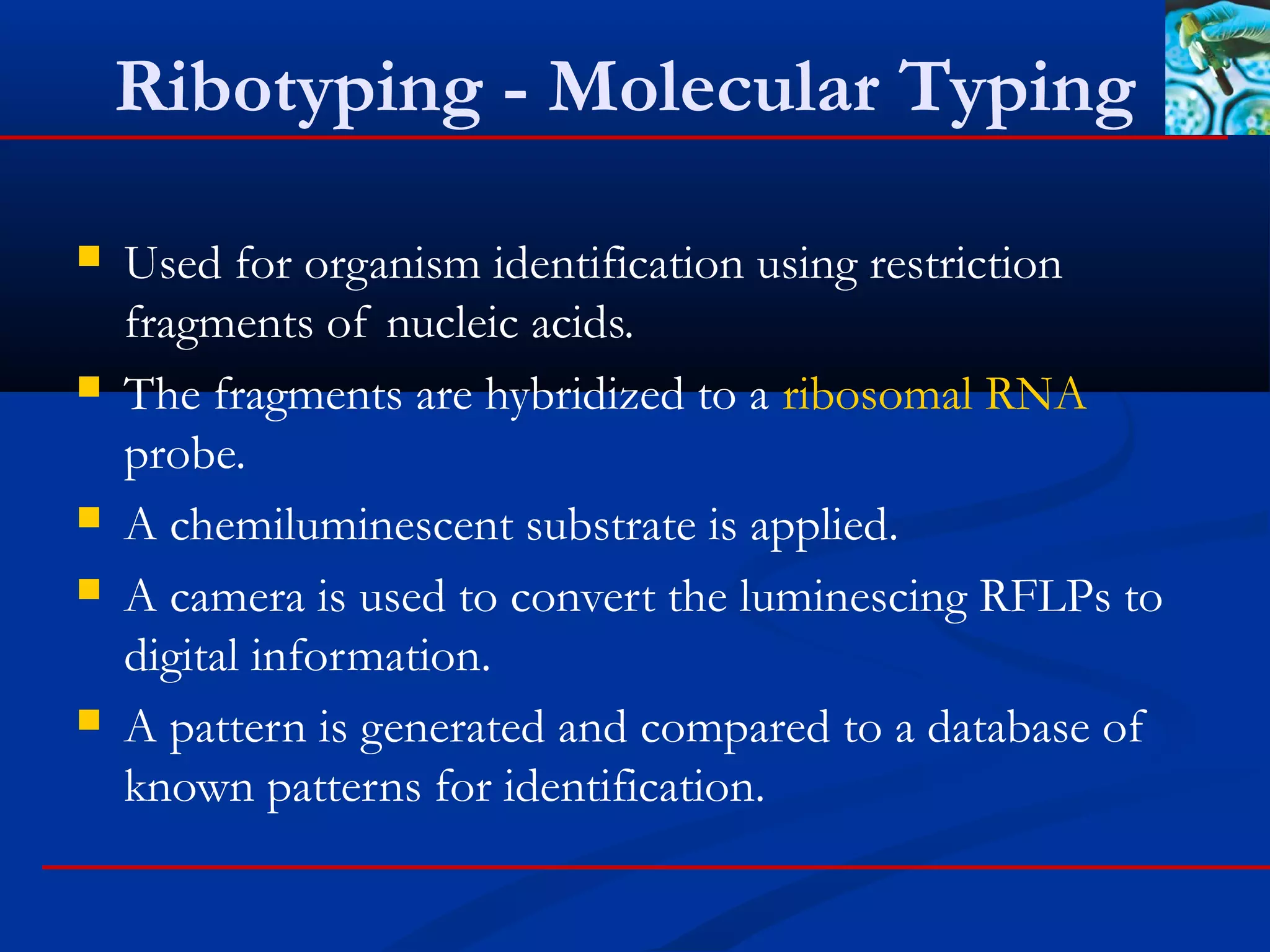
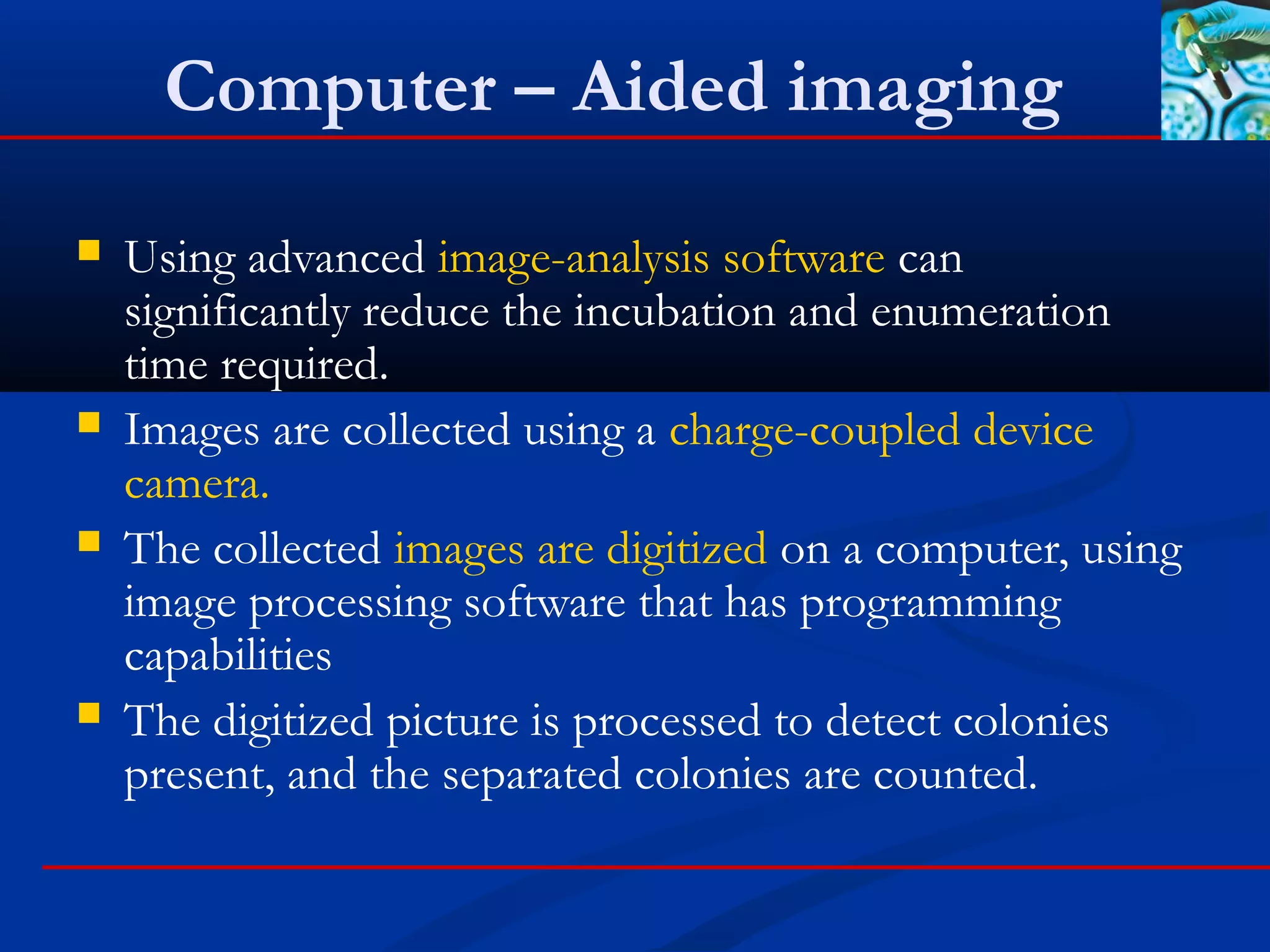
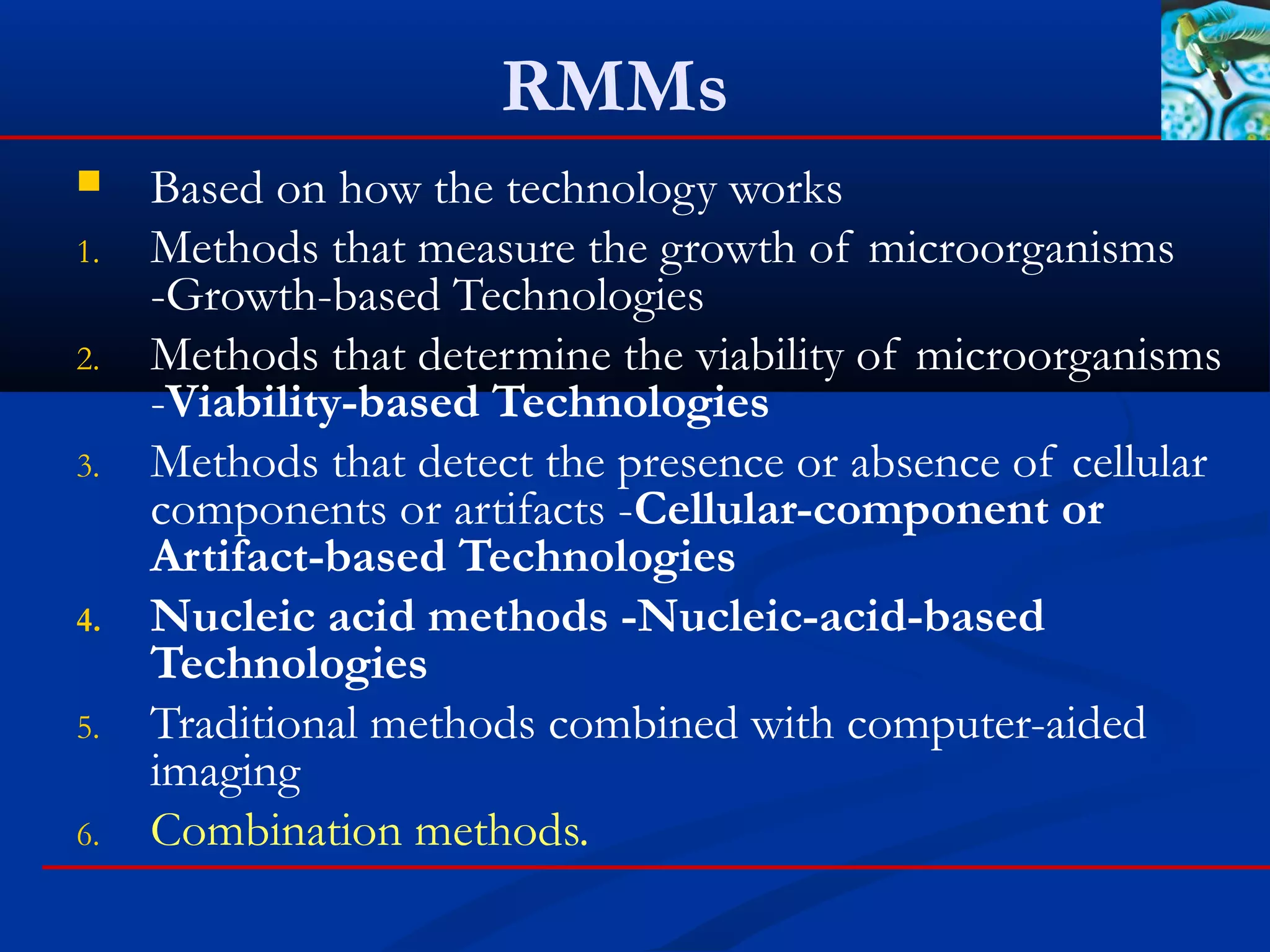
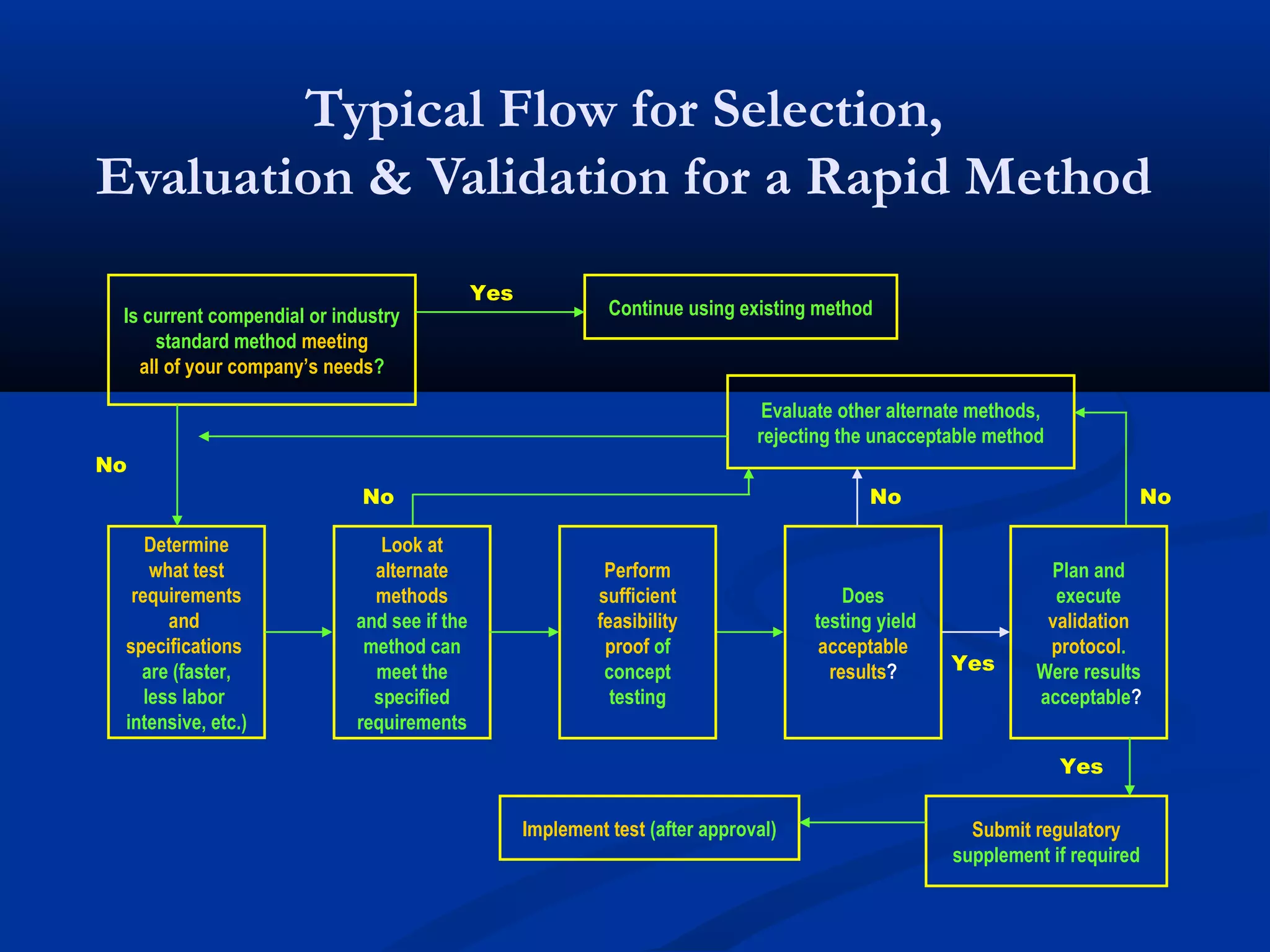
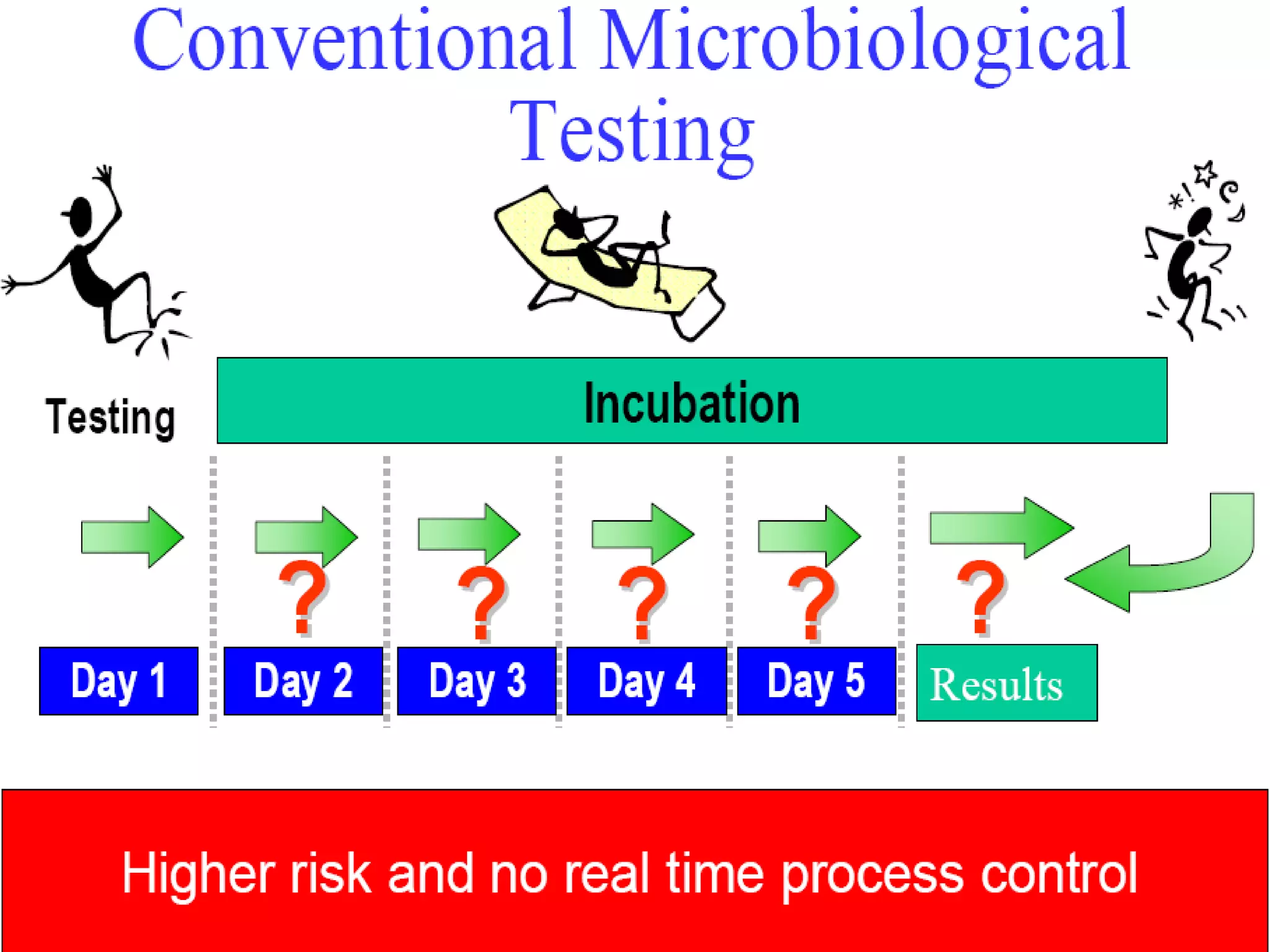
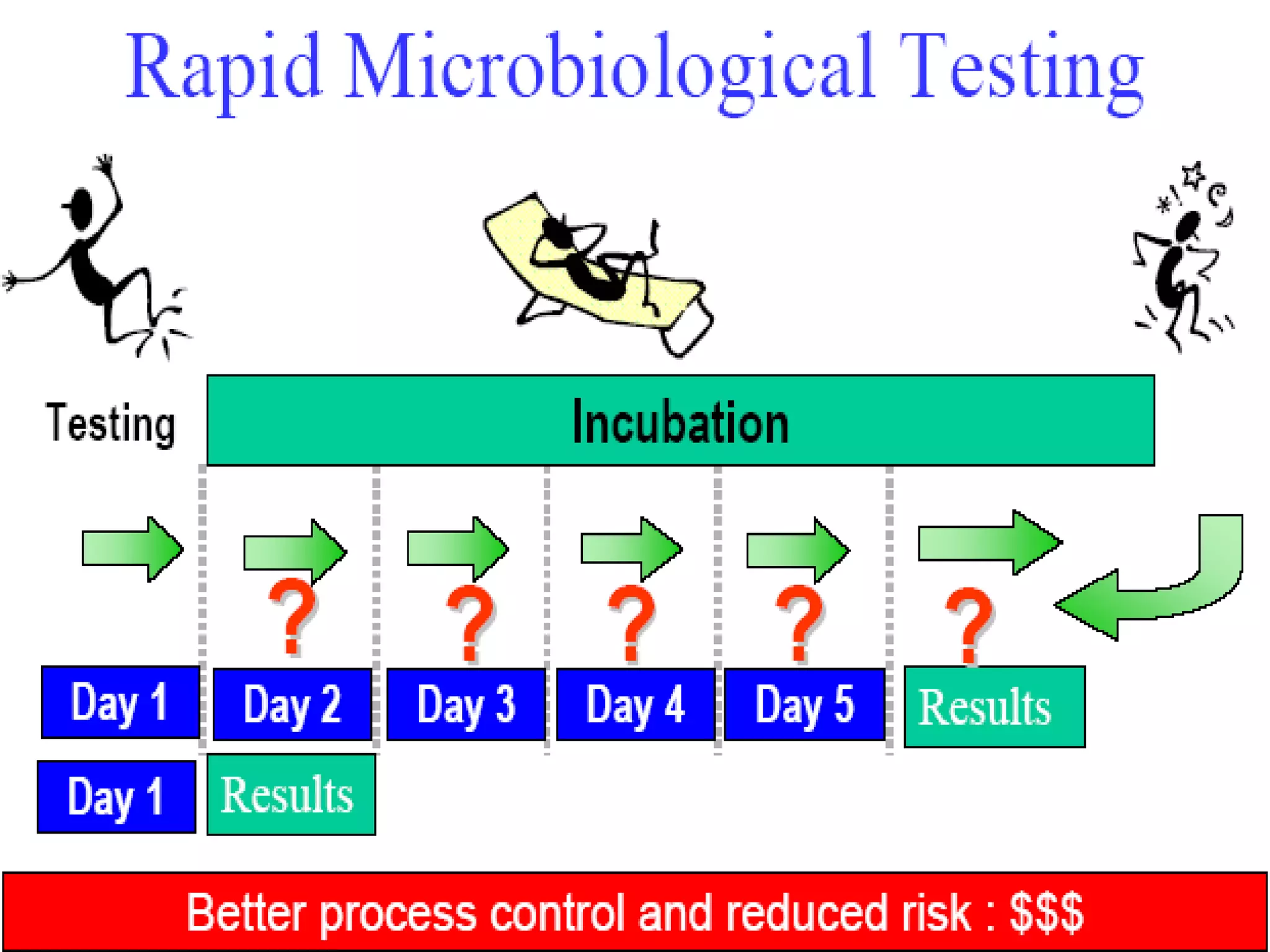
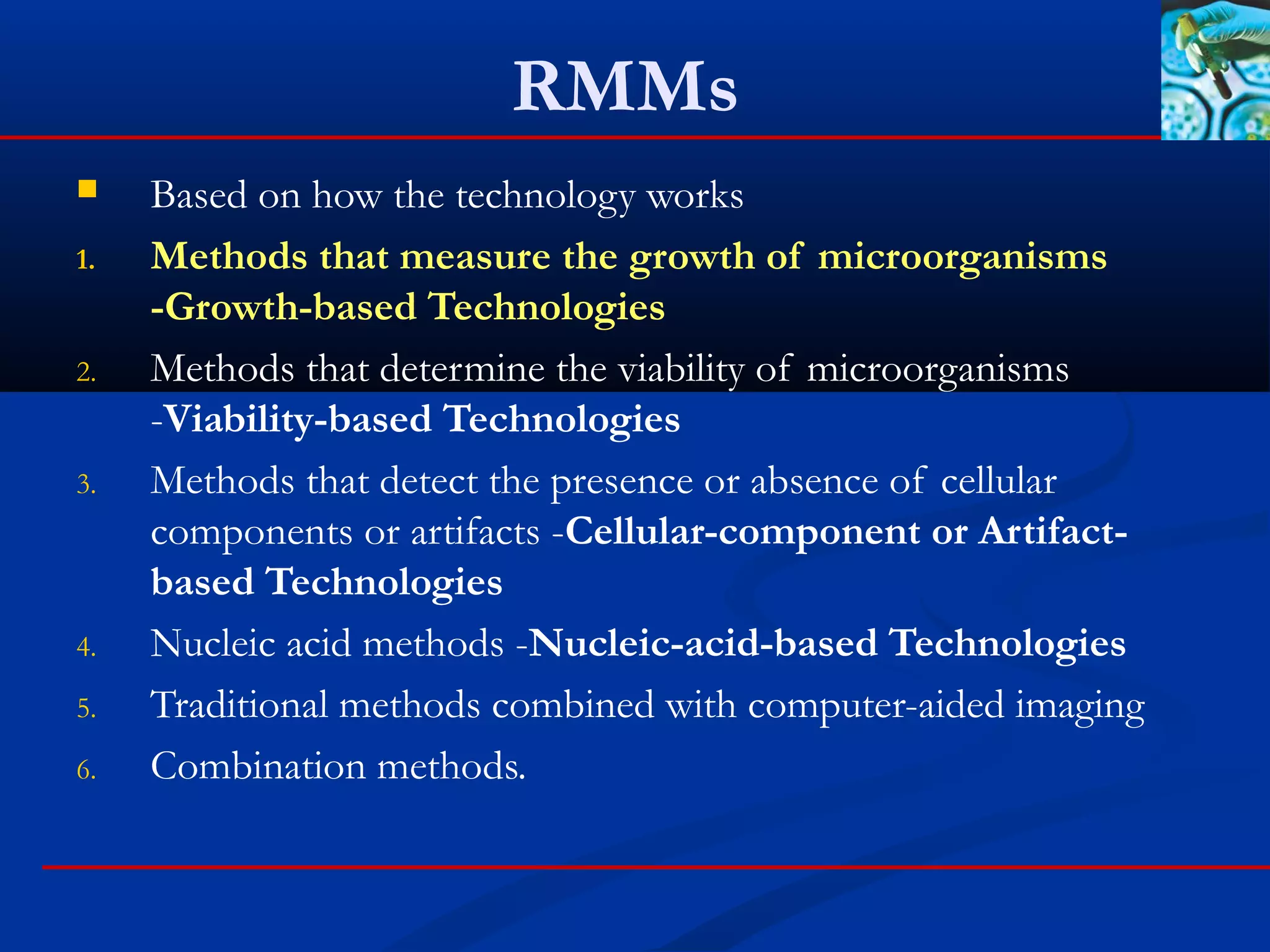
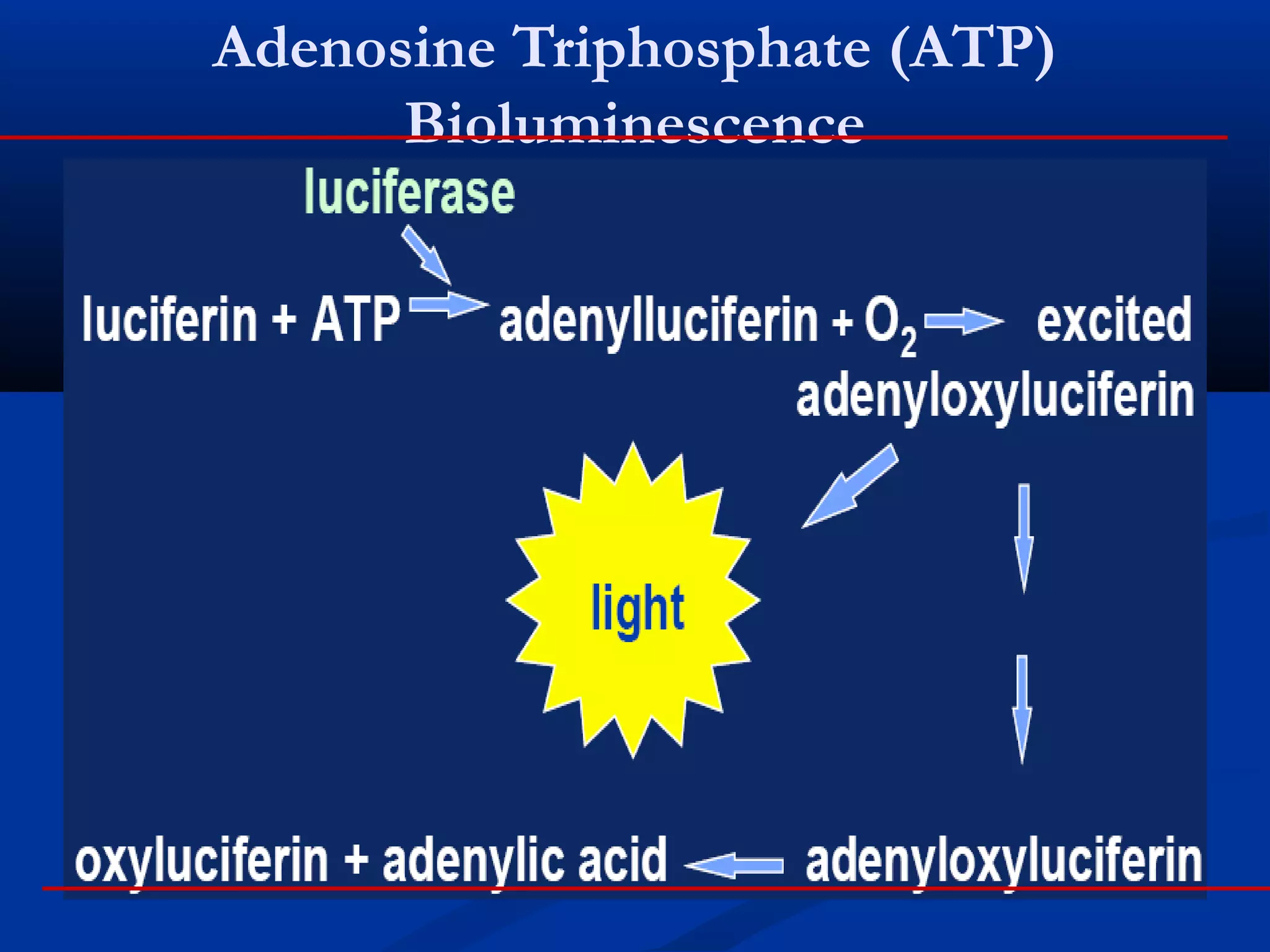
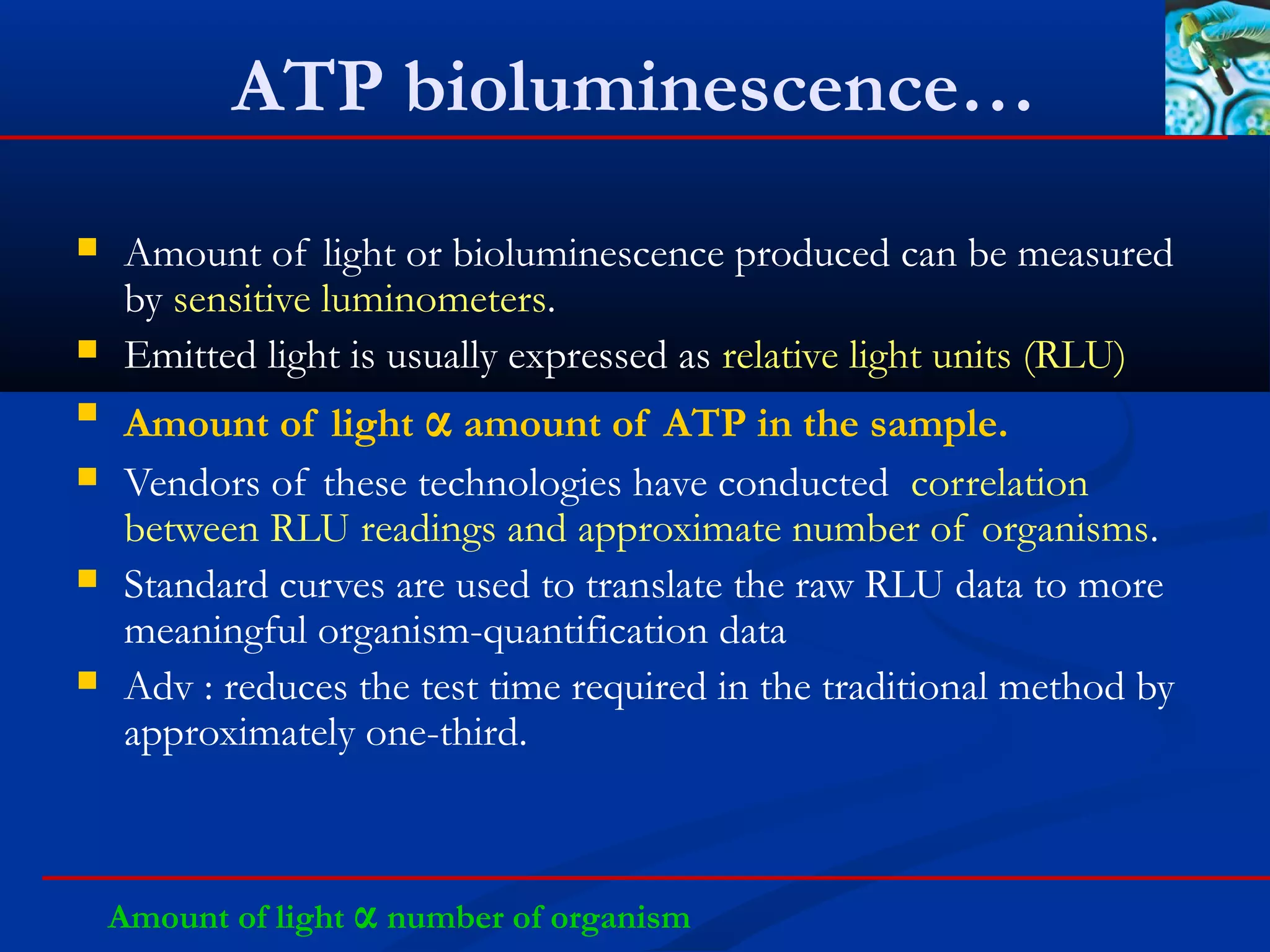
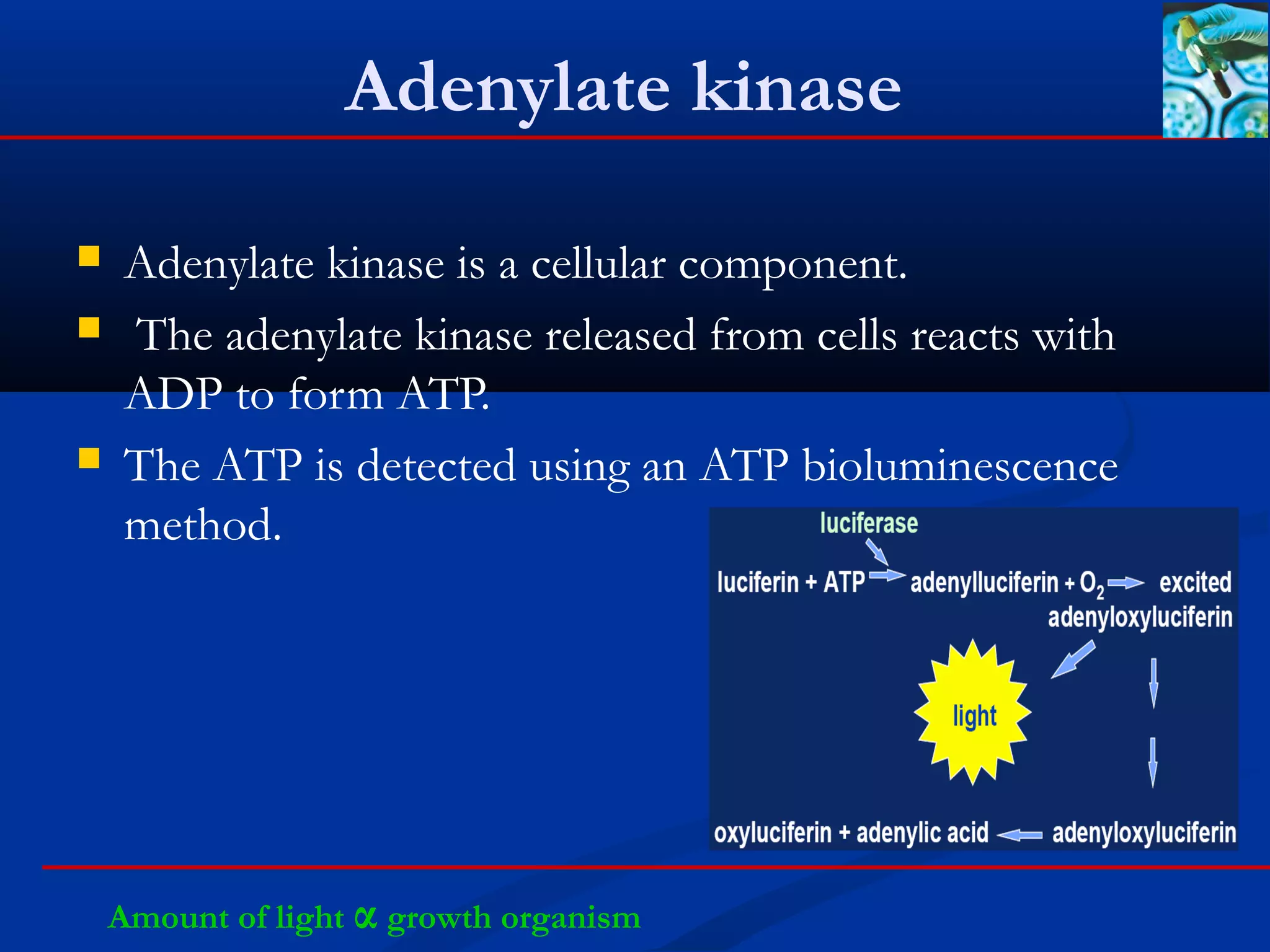
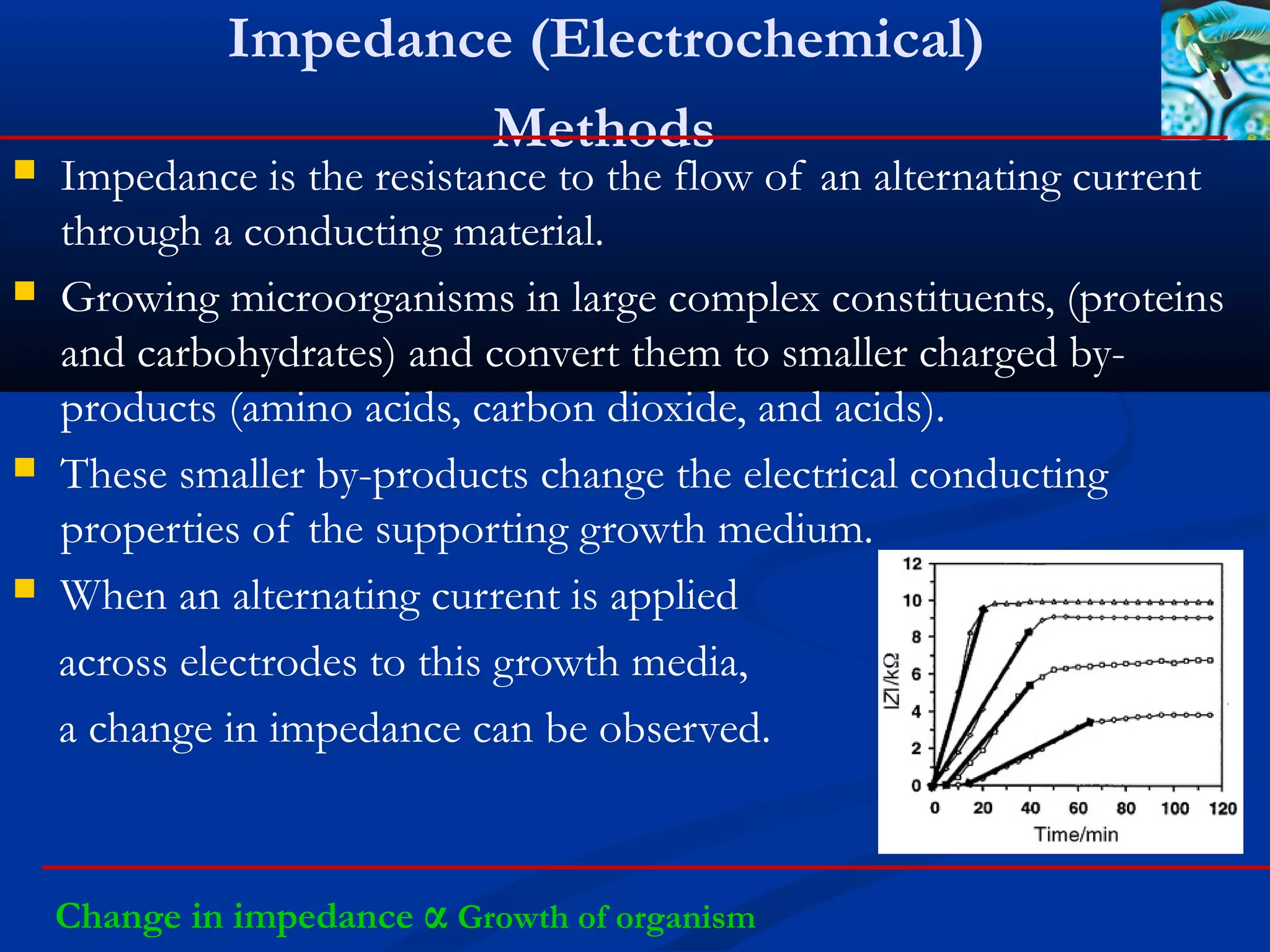
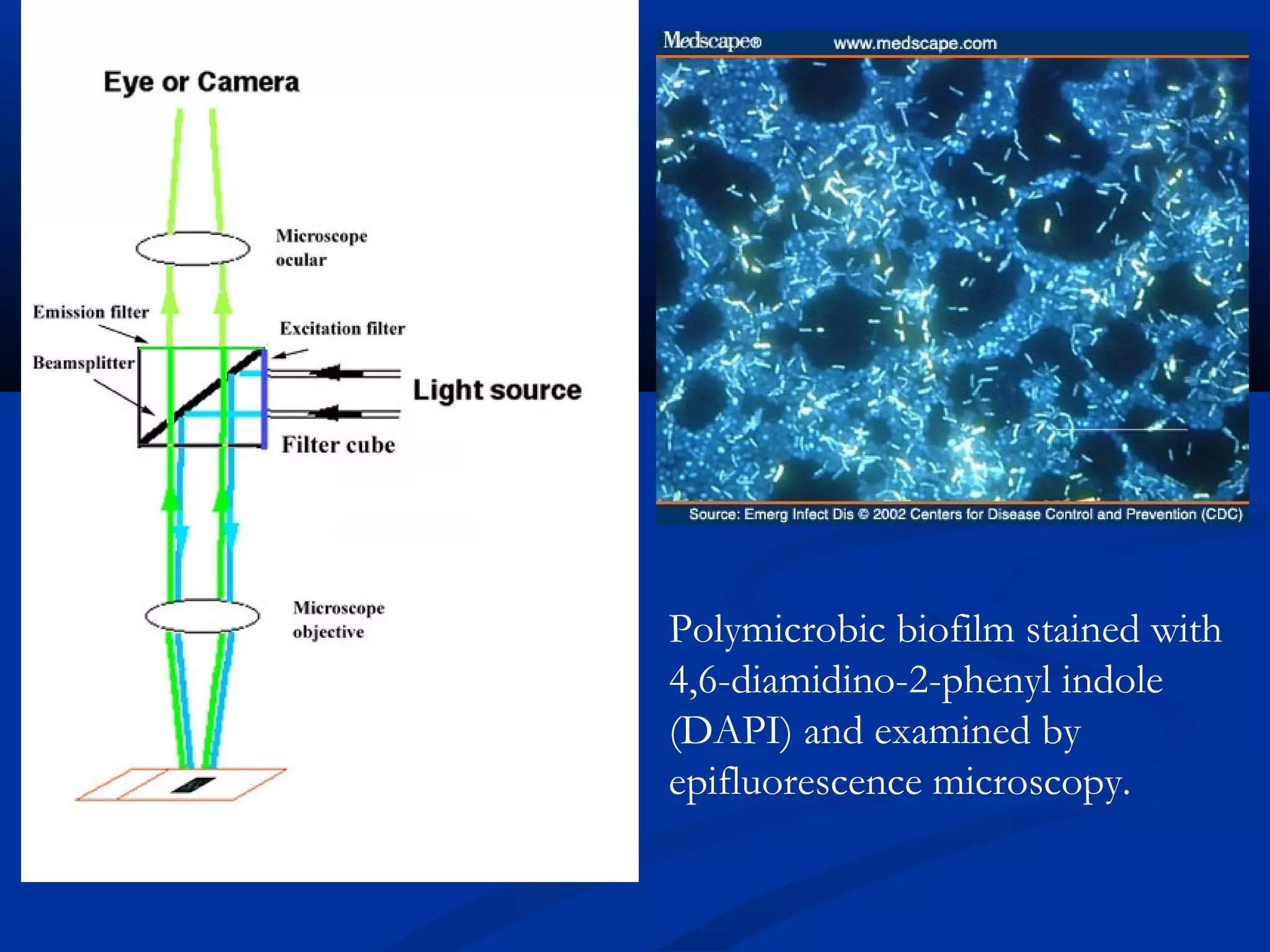
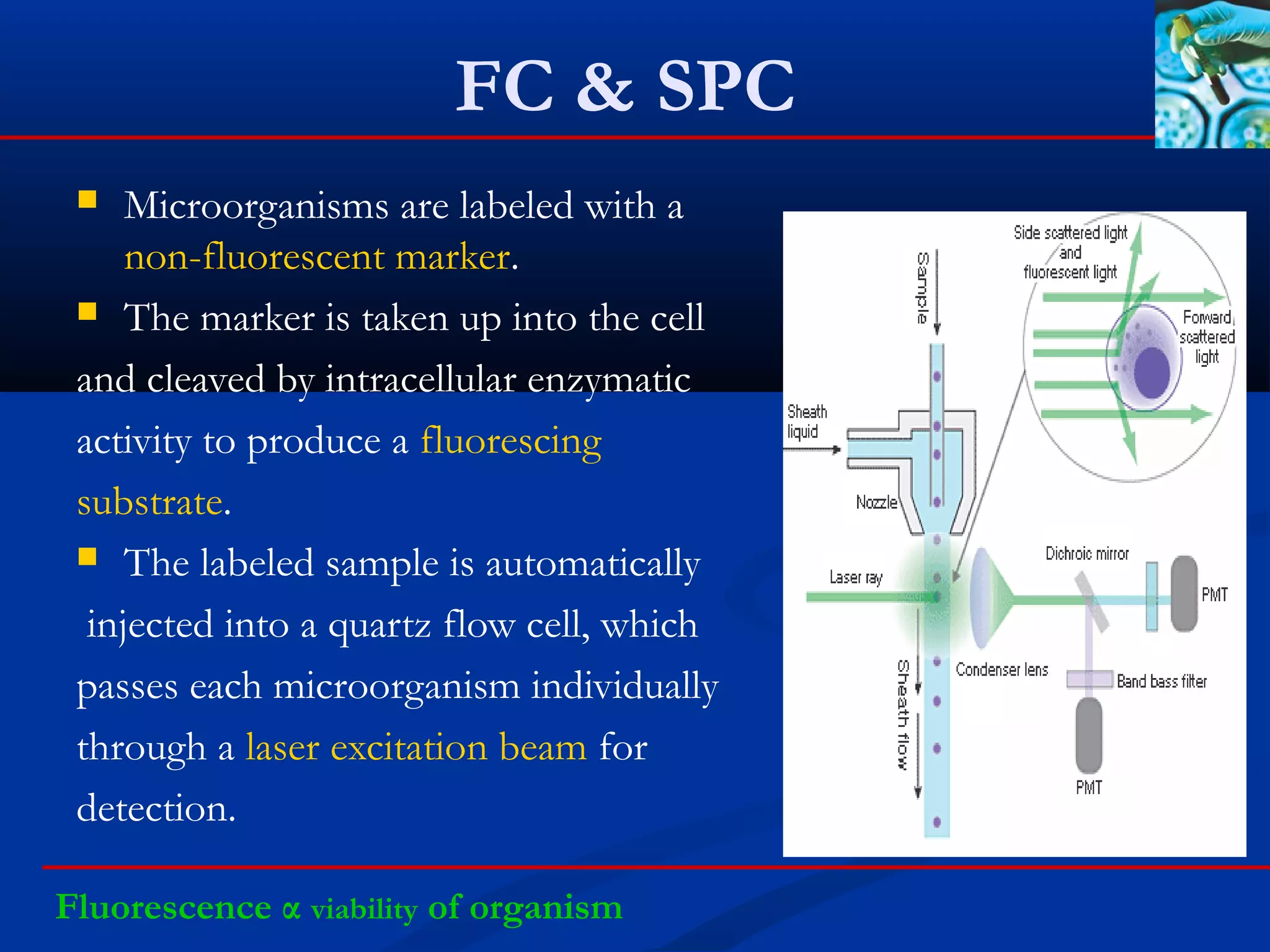
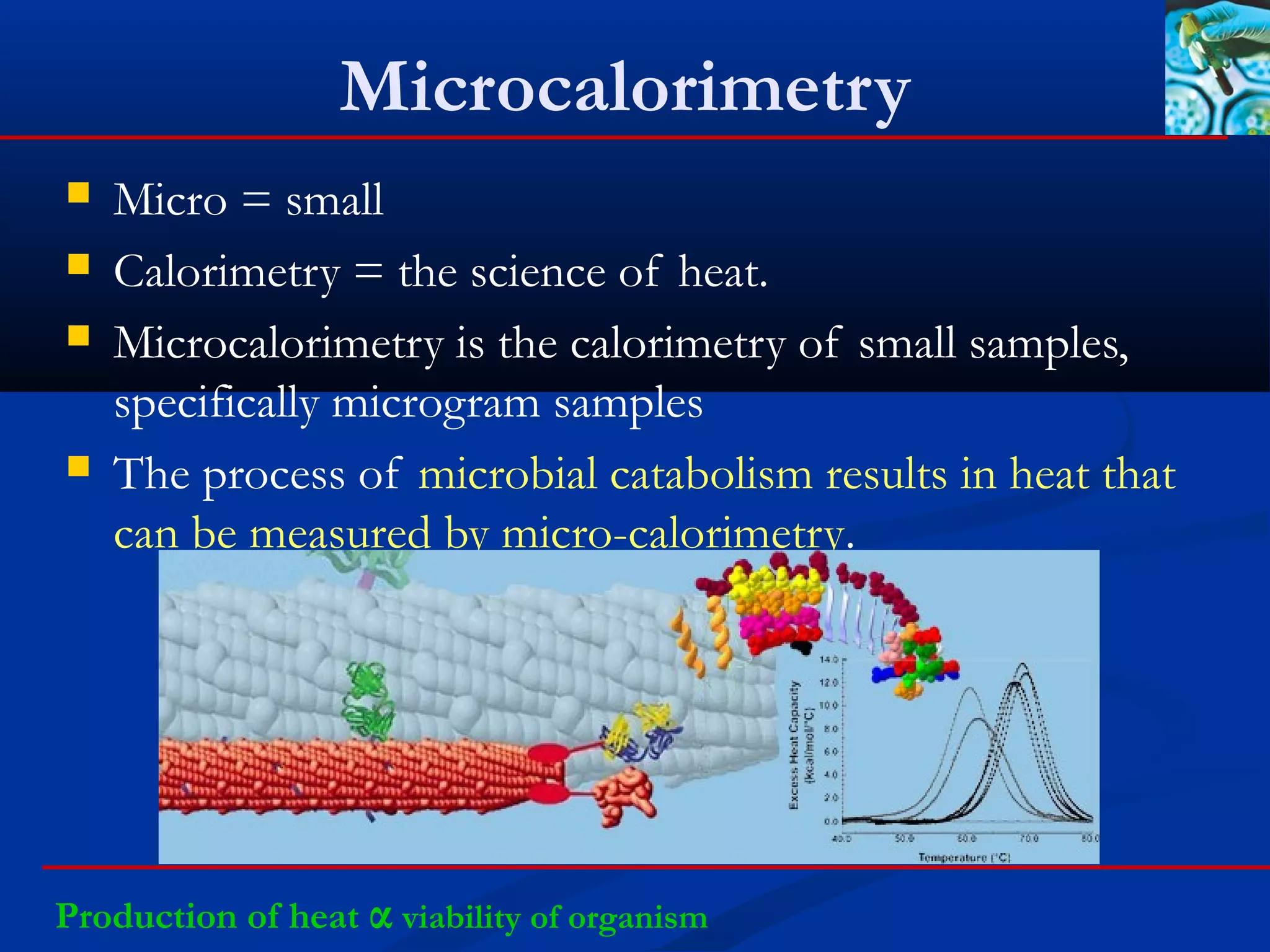
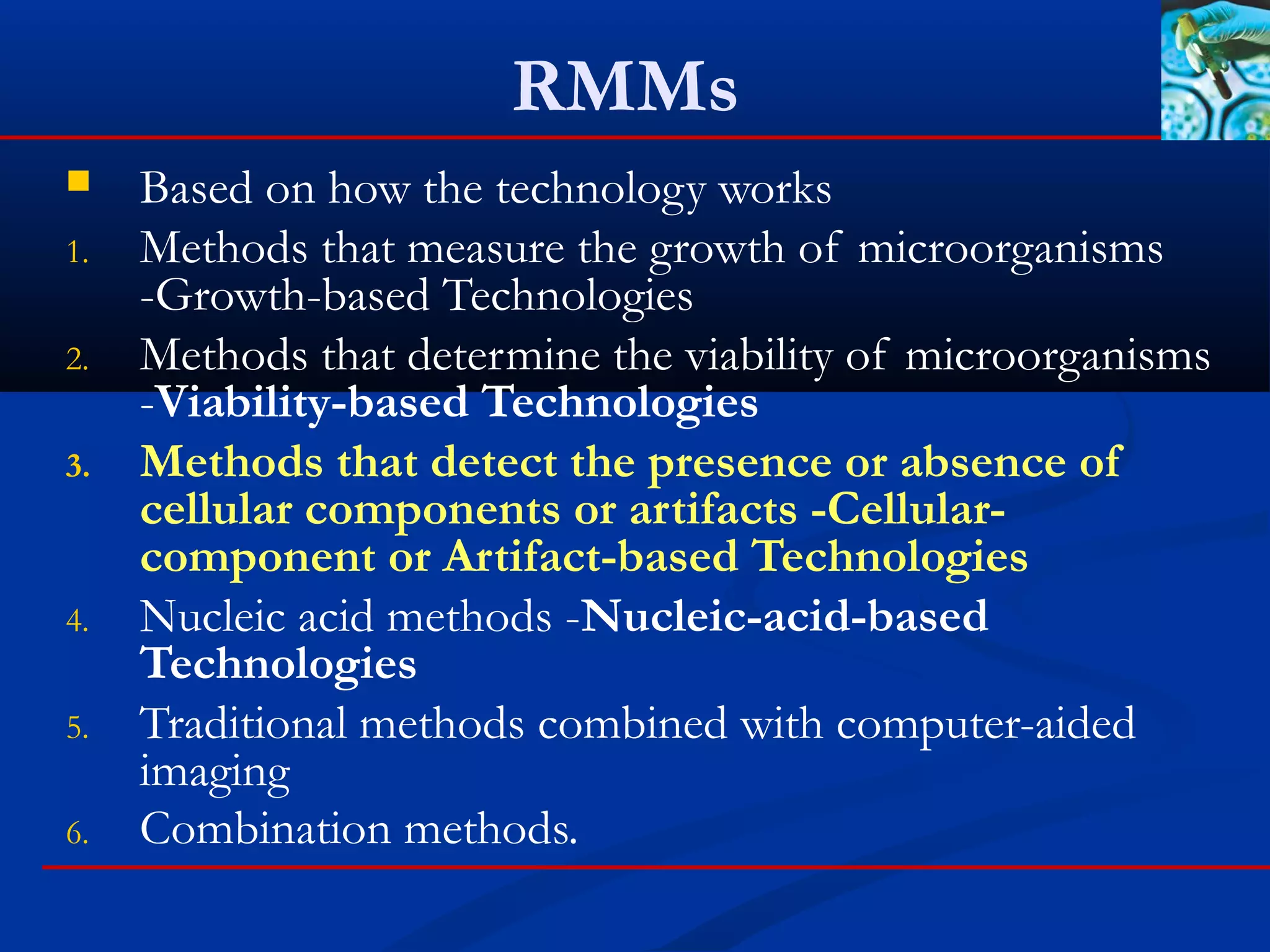
This document discusses rapid microbiological methods (RMMs) and the regulatory framework around validating and implementing new methods. It begins by explaining that traditional microbiological testing methods are slow and limited. RMMs aim to provide better, automated, and faster methods through technologies like ATP bioluminescence, nucleic acid detection, and computer-aided imaging. However, industries and regulators are slow to adopt new methods. The document then categorizes and provides examples of various RMM technologies before concluding that RMMs can provide advantages like speed, sensitivity, simplicity and reduced costs compared to traditional methods.











![Cellular-component based
Technologies
These technologies look for a specific cellular component or artifact
within the cell for detection or identification
Examples:
Fatty Acid Profiles (Fatty Acid Methyl Esters [FAMEs])
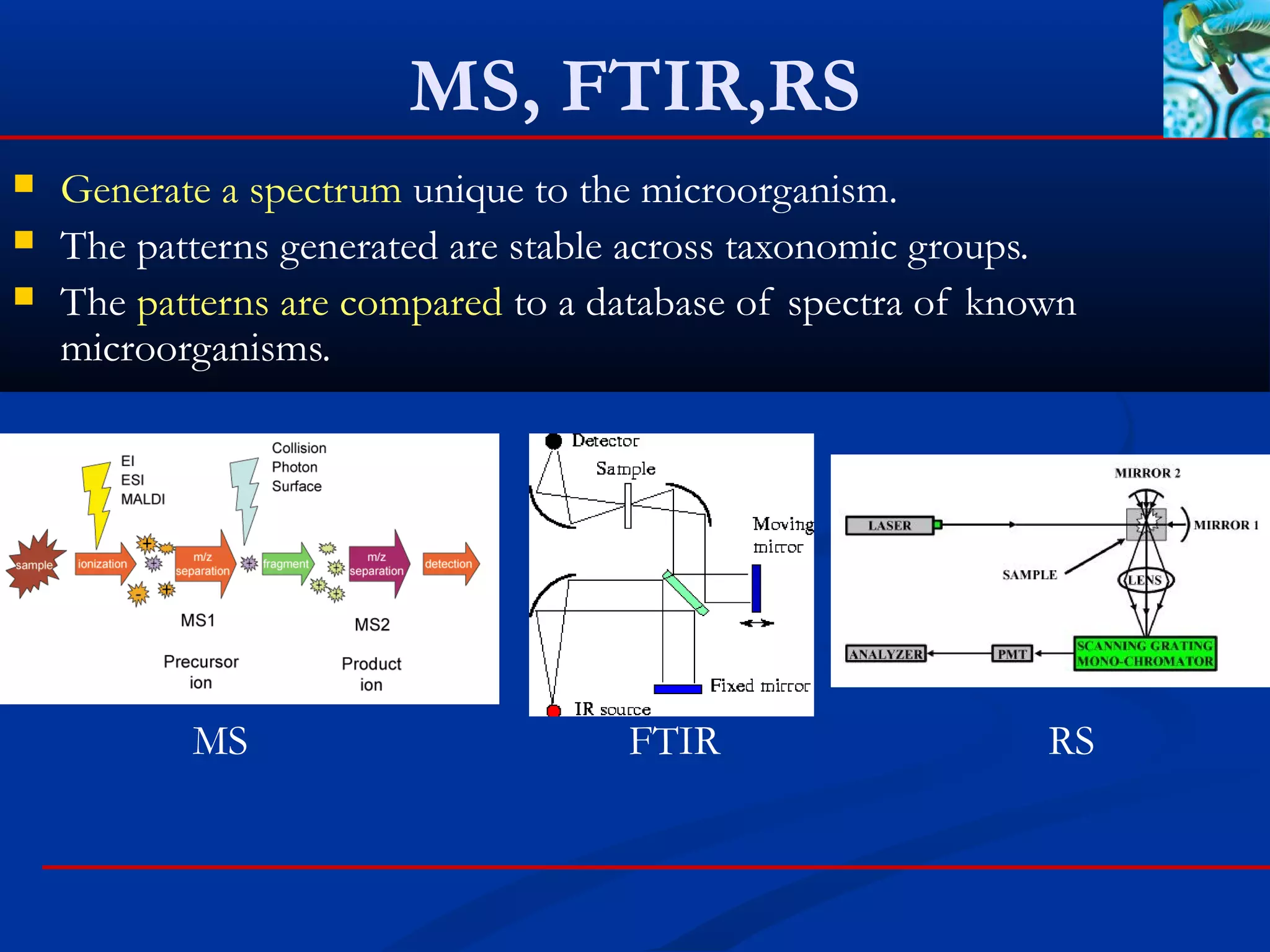
Mass spectrometry
Fourier Transform Infrared Spectroscopy (FTIR)
Raman spectroscopy
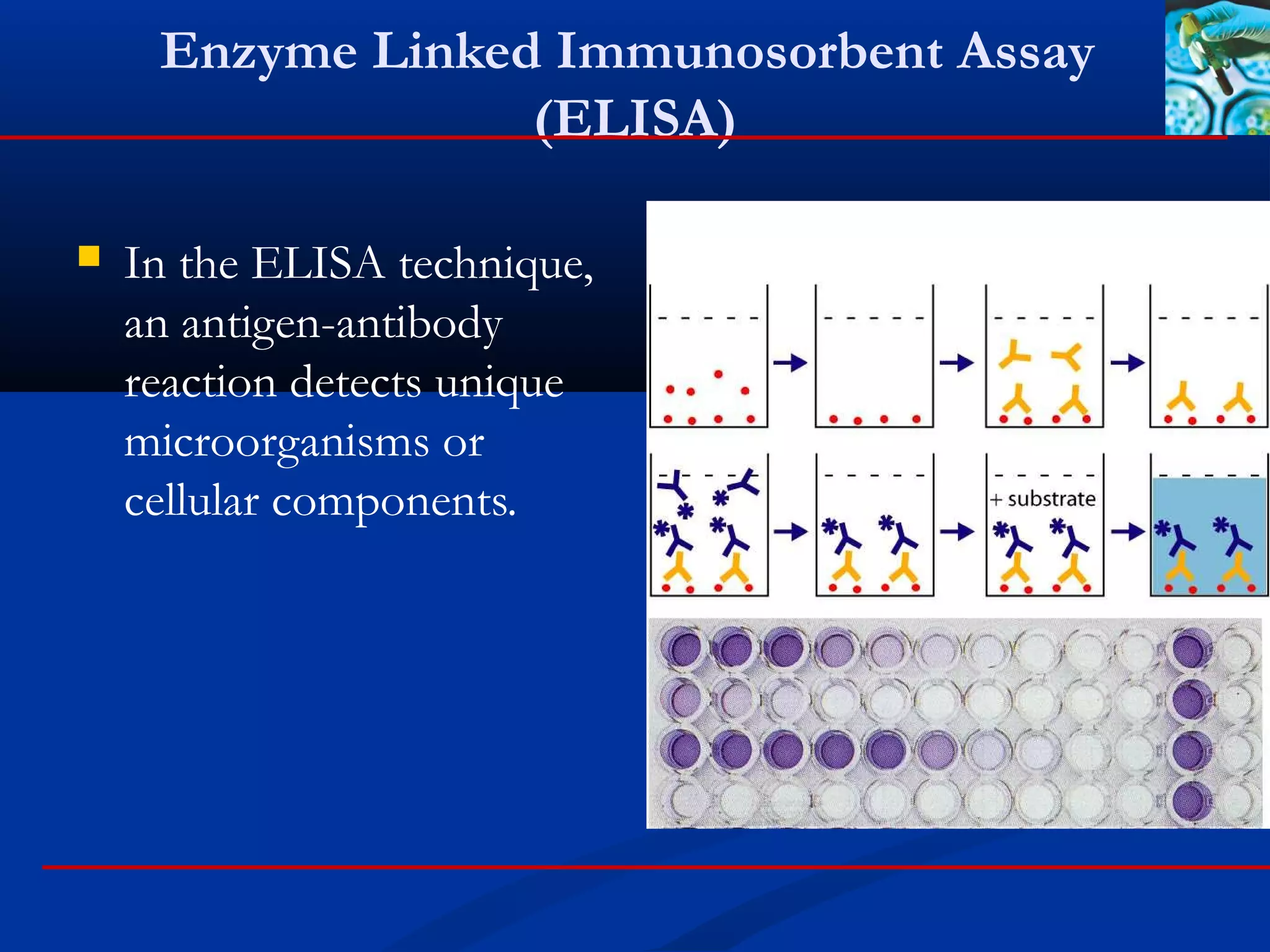
Enzyme linked immunosorbent assay (ELISA)
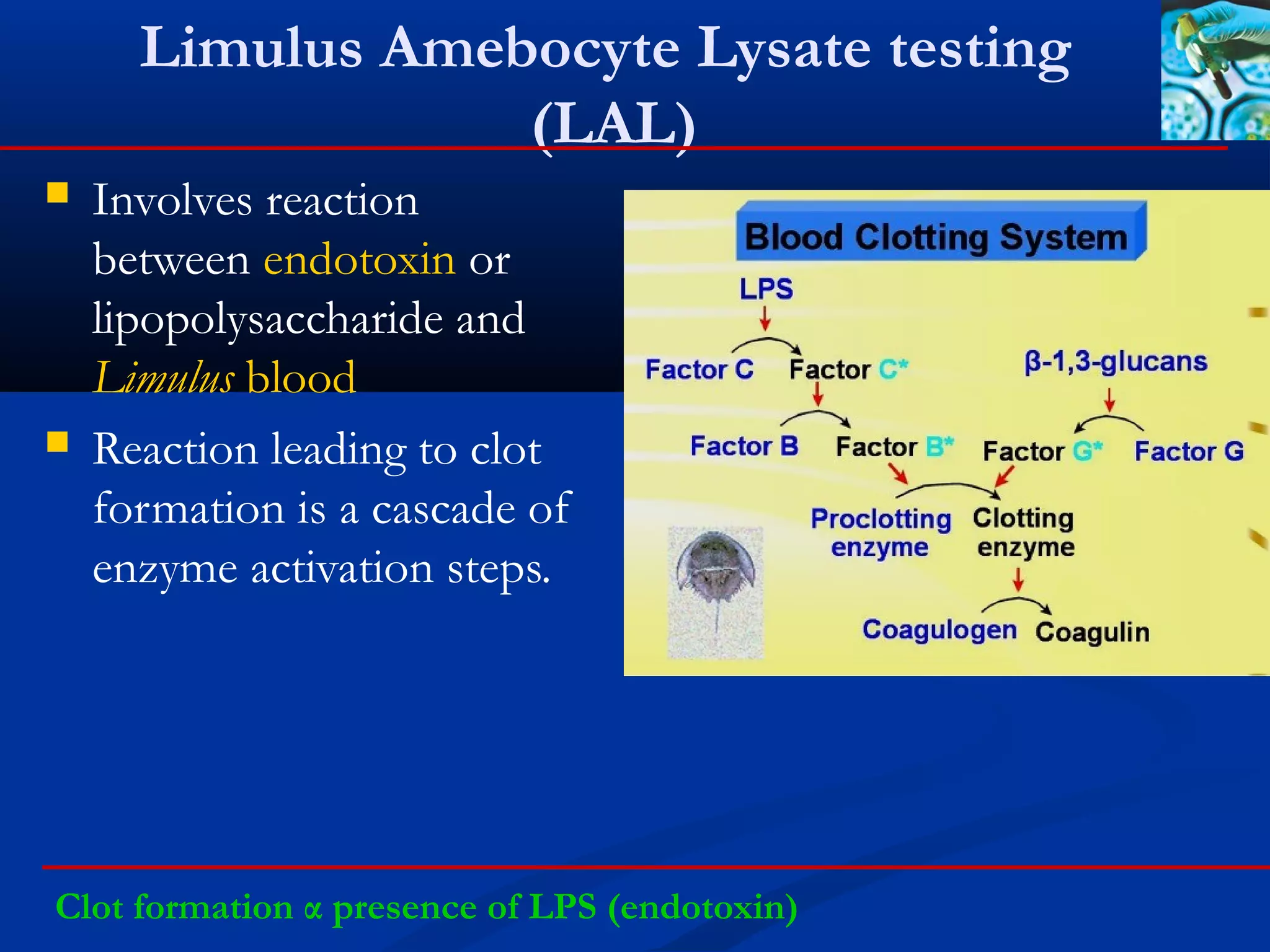
Bacterial endotoxin-limulus amebocyte lysate testing (LAL).
Endospore detection
Gram stains](https://image.slidesharecdn.com/rapidmicrobiologicalmethods-171115153450/75/Rapid-microbiological-methods-28-2048.jpg)
![Fatty Acid Profiles (Fatty Acid
Methyl Esters [FAMEs])
Fatty acids are present in
microorganisms
Isolates are grown on
standard media and selected
for testing.
The testing procedure
includes saponification of
fatty acids, methylation, and
extraction, to produce fatty
acid methyl esters (FAMEs).
The FAMEs are measured
using gas chromatography
Fatty acids → fatty acid methyl esters (FAMEs) → gas chromatography](https://image.slidesharecdn.com/rapidmicrobiologicalmethods-171115153450/75/Rapid-microbiological-methods-29-2048.jpg)
![Endospore detection
A major component of the spore case is calcium
dipicolinate (Ca[dpa]).
Dipicolinate anions (dpa2- ) are present only in bacterial
endospores.
Terbium (Tb3+ ) is able to complex with dpa2- , forming a
photoluminescence complex [Tb(dpa)]+
The sample is exposed to an ultraviolet (UV) source (250-
300 nm) and excited
dpa2- + Tb3+ →[Tb(dpa)]+ →ultraviolet source](https://image.slidesharecdn.com/rapidmicrobiologicalmethods-171115153450/75/Rapid-microbiological-methods-33-2048.jpg)