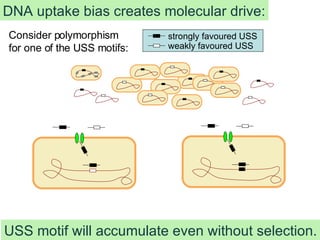
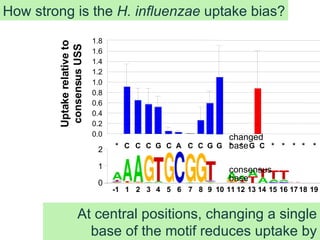
The talk by Rosie Redfield explores whether bacteria undergo processes akin to sexual reproduction, focusing on DNA uptake in Haemophilus influenzae. The study reveals that bacteria preferentially take up closely related DNA motifs that may not only serve nutritional purposes but also drive genomic change through biased DNA uptake. Ultimately, the findings suggest that this molecular drive can explain the accumulation of specific uptake sequence motifs in bacterial genomes.