- The document demonstrates various commands for exploring and summarizing data in R such as the iris data set including head(), tail(), str(), class(), summary(), and $-operator.
- The iris data set contains measurement data for 150 flowers across 4 variables and is stored as a data frame object in R.
- Data frames allow storing different data types together and can be explored using commands like summary() which provides summaries tailored to each variable type.
- Matrices can also be used to store multi-dimensional data and various functions like dim(), apply(), and cbind() allow manipulating the dimensions and combining matrices.
![Data available in R
> data()
> data("AirPassengers")
> head(AirPassengers)
[1] 112 118 132 129 121 135
> tail(AirPassengers)
[1] 622 606 508 461 390 432
> str(AirPassengers)
Time-Series [1:144] from 1949 to 1961: 112 118 132 129 121 135 148 148
136 119 ...
> class(AirPassengers)
[1] "ts"
> help(ts)
• The command data() loads data-sets available in R
• head() and tail() command displays first few or last few
values
• str() shows the structure of an R object
• class() shows the class of an R object
• What does “ts” stand for?](https://image.slidesharecdn.com/rpartiii-131204043801-phpapp01/75/R-part-iii-1-2048.jpg)
![Try runif() and plot() commands ….
runif(10)
[1] 0.14350413 0.54293576 0.62881627 0.30278850 0.28030129 0.03784996
0.49483957
[8] 0.23571517 0.40072956 0.20327478
> plot(runif(10))
The runif()
command generates
U(0,1)10 random
numbers between 0
and 1.
These numbers have
been plotted by the
plot() function.](https://image.slidesharecdn.com/rpartiii-131204043801-phpapp01/75/R-part-iii-2-2048.jpg)
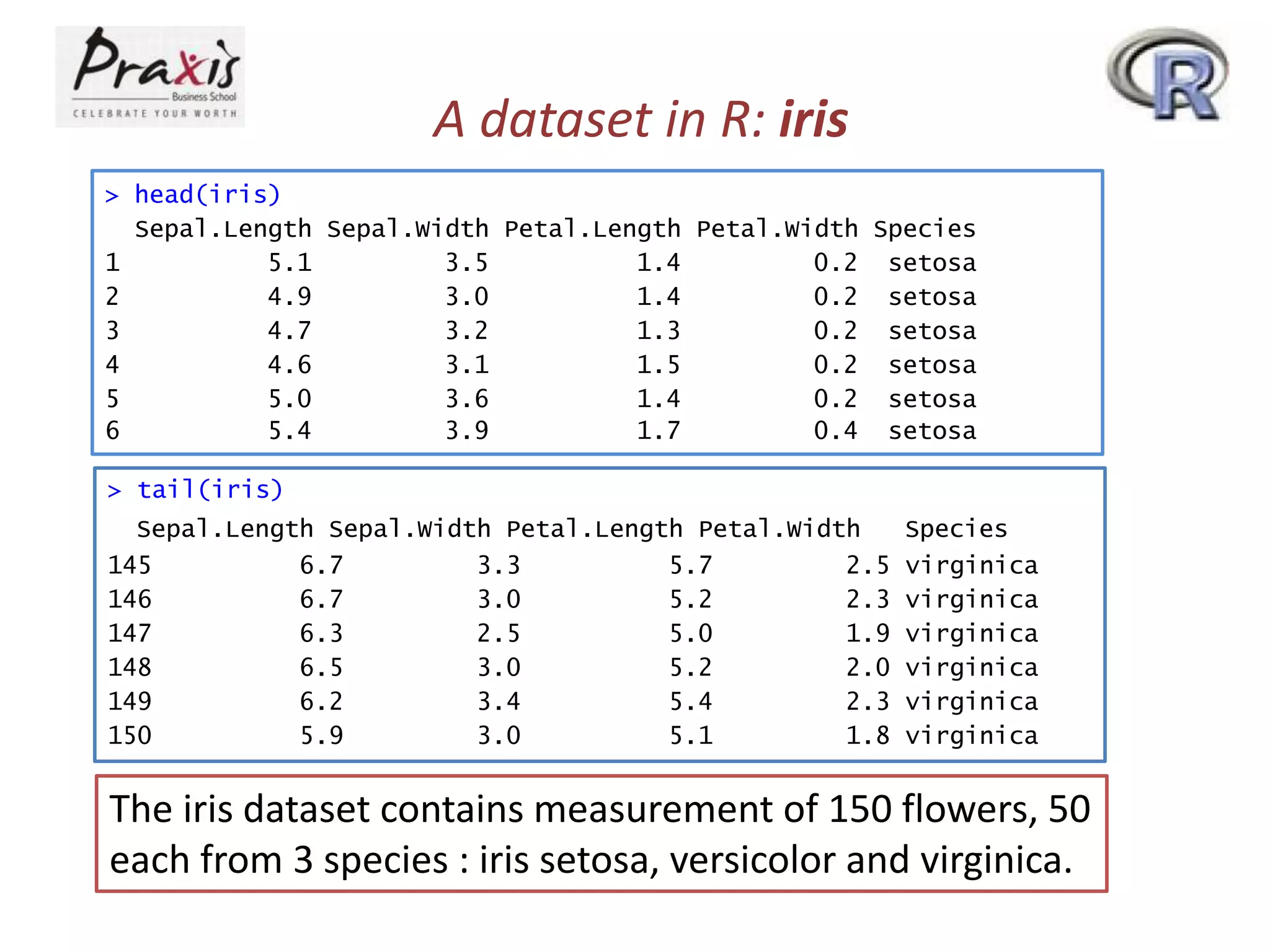
![Data frame in R: iris
> str(iris)
'data.frame':
150 obs. of 5 variables:
$ Sepal.Length: num 5.1 4.9 4.7 4.6 5 5.4 4.6 5 4.4 4.9 ...
$ Sepal.Width : num 3.5 3 3.2 3.1 3.6 3.9 3.4 3.4 2.9 3.1 ...
$ Petal.Length: num 1.4 1.4 1.3 1.5 1.4 1.7 1.4 1.5 1.4 1.5 ...
$ Petal.Width : num 0.2 0.2 0.2 0.2 0.2 0.4 0.3 0.2 0.2 0.1 ...
$ Species
: Factor w/ 3 levels "setosa","versicolor",..: 1 1 1 1 1
1 1 1 1 1 ...
> class(iris)
[1] "data.frame"
• As you see, iris is not a simple vector but a composite
“data frame” object made up of several component
vectors as you can see in the output of class(iris)
• You can think of a data frame as a matrix-like object
- each row for each observational unit (here, a flower)
- each column for each measurement made on the unit
• But the str() function gives you more concise description
on iris.](https://image.slidesharecdn.com/rpartiii-131204043801-phpapp01/75/R-part-iii-4-2048.jpg)
![Use of $ operator: iris
> iris$Sepal.Length
[1] 5.1 4.9 4.7 4.6
5.7 5.1
[21] 5.4 5.1 4.6 5.1
4.4 5.1
[41] 5.0 4.5 4.4 5.0
6.6 5.2
[61] 5.0 5.9 6.0 6.1
6.0 5.7
[81] 5.5 5.5 5.8 6.0
5.1 5.7
[101] 6.3 5.8 7.1 6.3
7.7 6.0
[121] 6.9 5.6 7.7 6.3
6.0 6.9
[141] 6.7 6.9 5.8 6.8
5.0 5.4 4.6 5.0 4.4 4.9 5.4 4.8 4.8 4.3 5.8 5.7 5.4 5.1
4.8 5.0 5.0 5.2 5.2 4.7 4.8 5.4 5.2 5.5 4.9 5.0 5.5 4.9
5.1 4.8 5.1 4.6 5.3 5.0 7.0 6.4 6.9 5.5 6.5 5.7 6.3 4.9
5.6 6.7 5.6 5.8 6.2 5.6 5.9 6.1 6.3 6.1 6.4 6.6 6.8 6.7
5.4 6.0 6.7 6.3 5.6 5.5 5.5 6.1 5.8 5.0 5.6 5.7 5.7 6.2
6.5 7.6 4.9 7.3 6.7 7.2 6.5 6.4 6.8 5.7 5.8 6.4 6.5 7.7
6.7 7.2 6.2 6.1 6.4 7.2 7.4 7.9 6.4 6.3 6.1 7.7 6.3 6.4
6.7 6.7 6.3 6.5 6.2 5.9
Note that $-operator extracts individual components of a data
frame.
Try summary() and IQR() commands on iris$Sepal.Length
and study the data](https://image.slidesharecdn.com/rpartiii-131204043801-phpapp01/75/R-part-iii-5-2048.jpg)
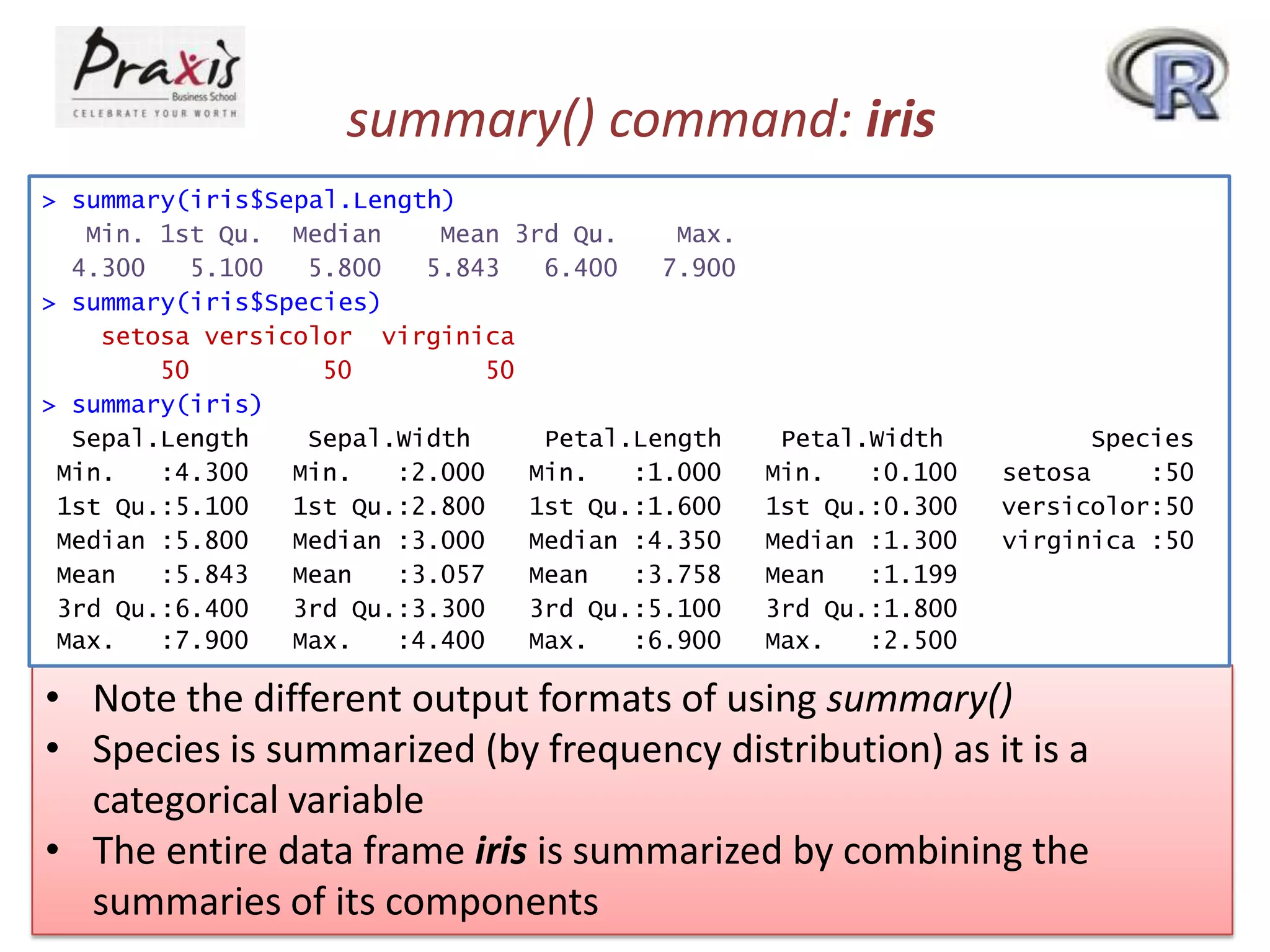
![class() command: iris
> class(iris$Sepal.Length)
[1] "numeric"
> class(iris$Species)
[1] "factor"
> class(iris)
[1] "data.frame"
• Note that each R object has a class (“numeric”, “factor” etc.)
• summary() is referred to as a generic function
• When summary() is applied, R figures out the appropriate
method and calls it](https://image.slidesharecdn.com/rpartiii-131204043801-phpapp01/75/R-part-iii-7-2048.jpg)
![More on summary() command
> methods(summary)
[1] summary.aov
[4] summary.connection
[7] summary.default
[10] summary.glm
[13] summary.loess*
[16] summary.mlm
[19] summary.PDF_Dictionary*
[22] summary.POSIXlt
[25] summary.princomp*
[28] summary.stepfun
[31] summary.tukeysmooth*
summary.aovlist
summary.data.frame
summary.ecdf*
summary.infl
summary.manova
summary.nls*
summary.PDF_Stream*
summary.ppr*
summary.srcfile
summary.stl*
summary.aspell*
summary.Date
summary.factor
summary.lm
summary.matrix
summary.packageStatus*
summary.POSIXct
summary.prcomp*
summary.srcref
summary.table
Non-visible functions are asterisked
• Objects of class “factor” are handled by summary.factor()
• “data.frame”s are handled by summary.data.frame()
• Numeric vectors are handled by summary.default()](https://image.slidesharecdn.com/rpartiii-131204043801-phpapp01/75/R-part-iii-8-2048.jpg)
![Try the following ….
•
•
•
•
•
•
•
•
•
attach() and detach() with iris
xx <- 1:12 and then dim(xx) <- c(3,4)
apply nrow(xx) and ncol(xx)
dim(xx) <- c(2,2,3)
yy <- matrix(1:12, nrows=3, byrow=TRUE
rownames(yy) <- LETTERS[1:3]
use colnames()
zz <- cbind(A=1:4, B=5:8, C=9:12)
rbind(zz,0)](https://image.slidesharecdn.com/rpartiii-131204043801-phpapp01/75/R-part-iii-9-2048.jpg)