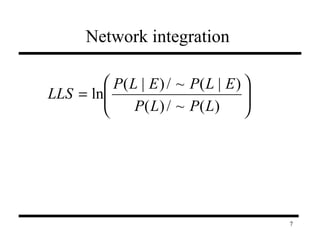
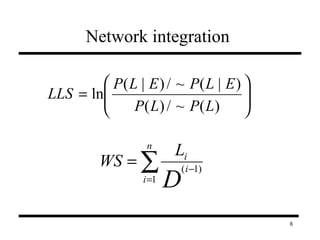
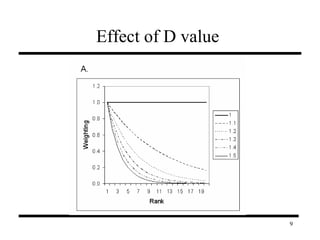
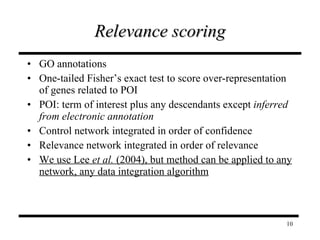
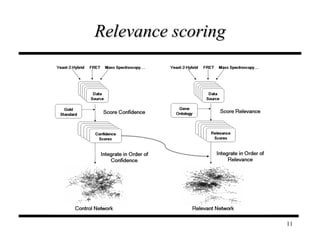
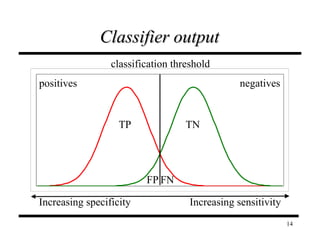
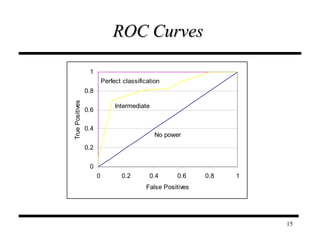
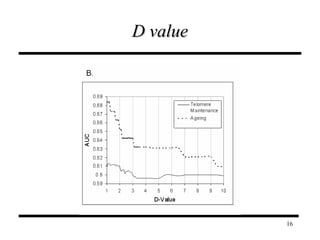
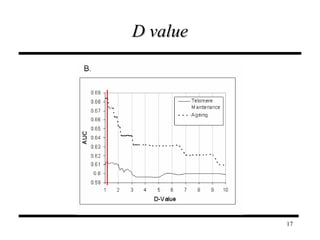
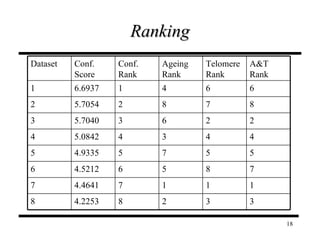
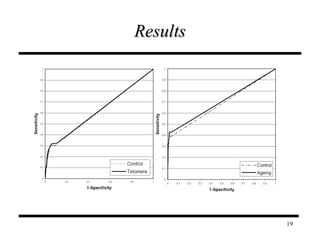
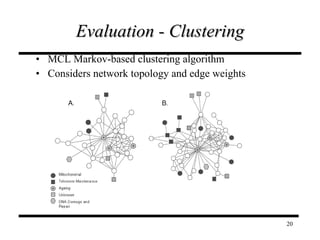
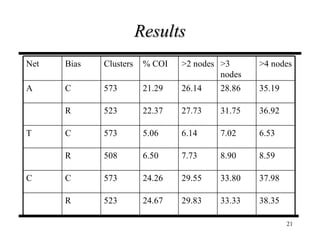
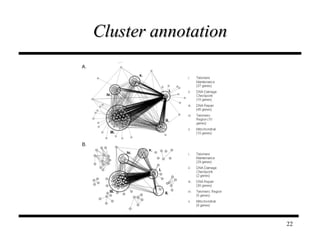
The document discusses integrating probabilistic functional networks to identify interactions relevant to specific biological processes. It introduces a method to score interactions based on their relevance to a process of interest using gene ontology annotations. The method is tested on Saccharomyces cerevisiae protein interaction data to build networks relevant to telomere maintenance and aging. Evaluation shows relevance networks contain more clustered genes of interest compared to networks built only on interaction confidence.