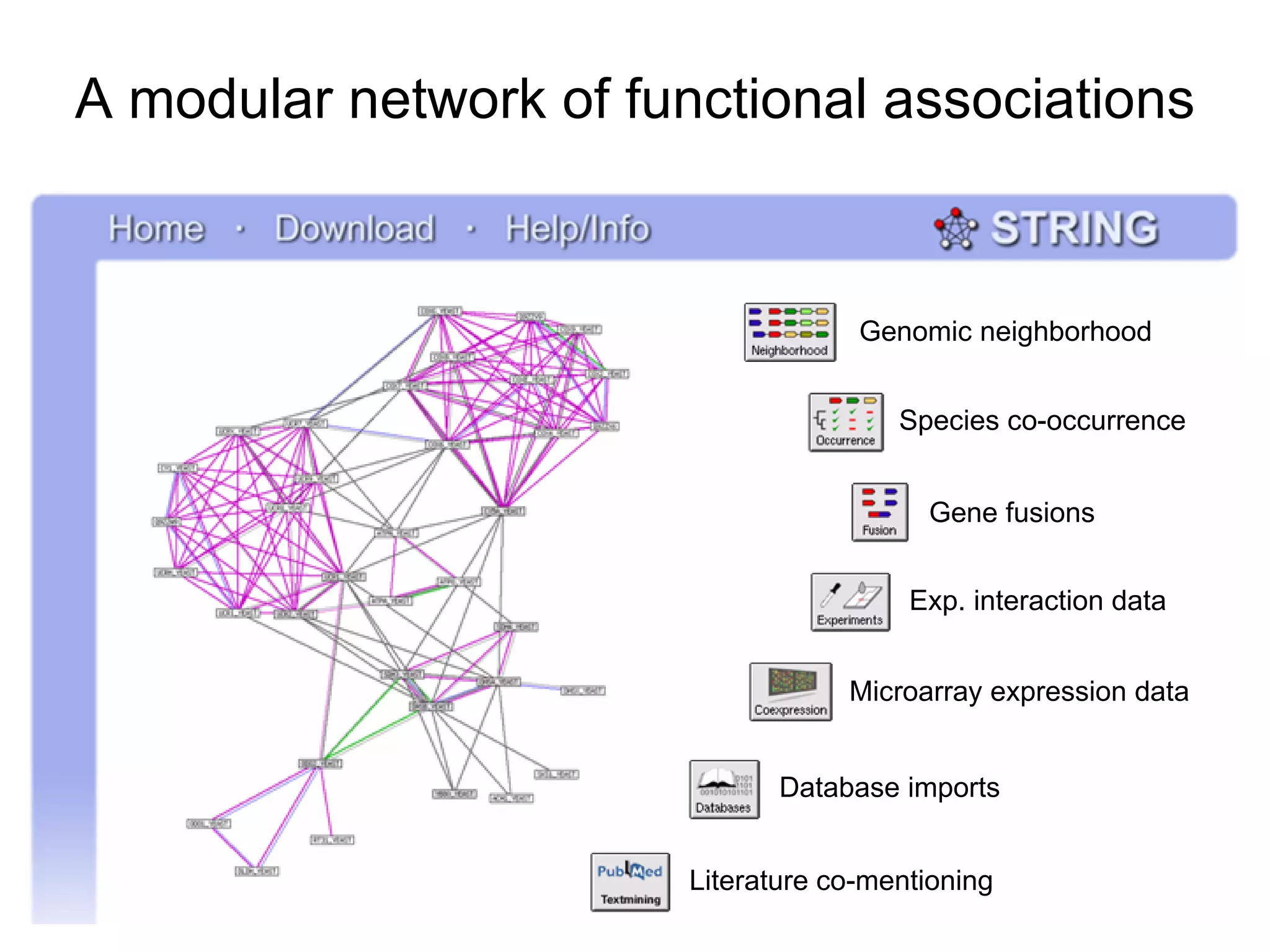
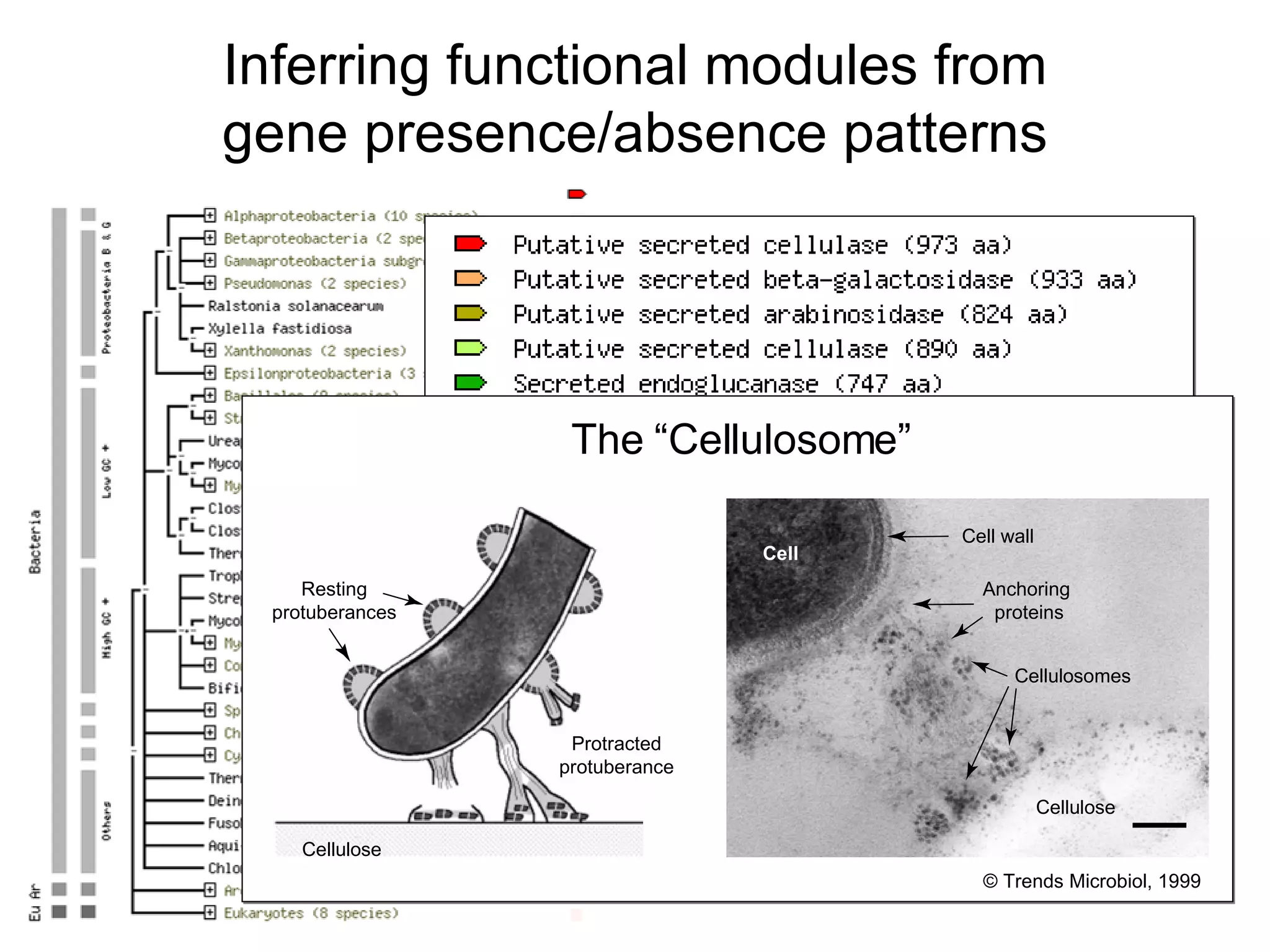
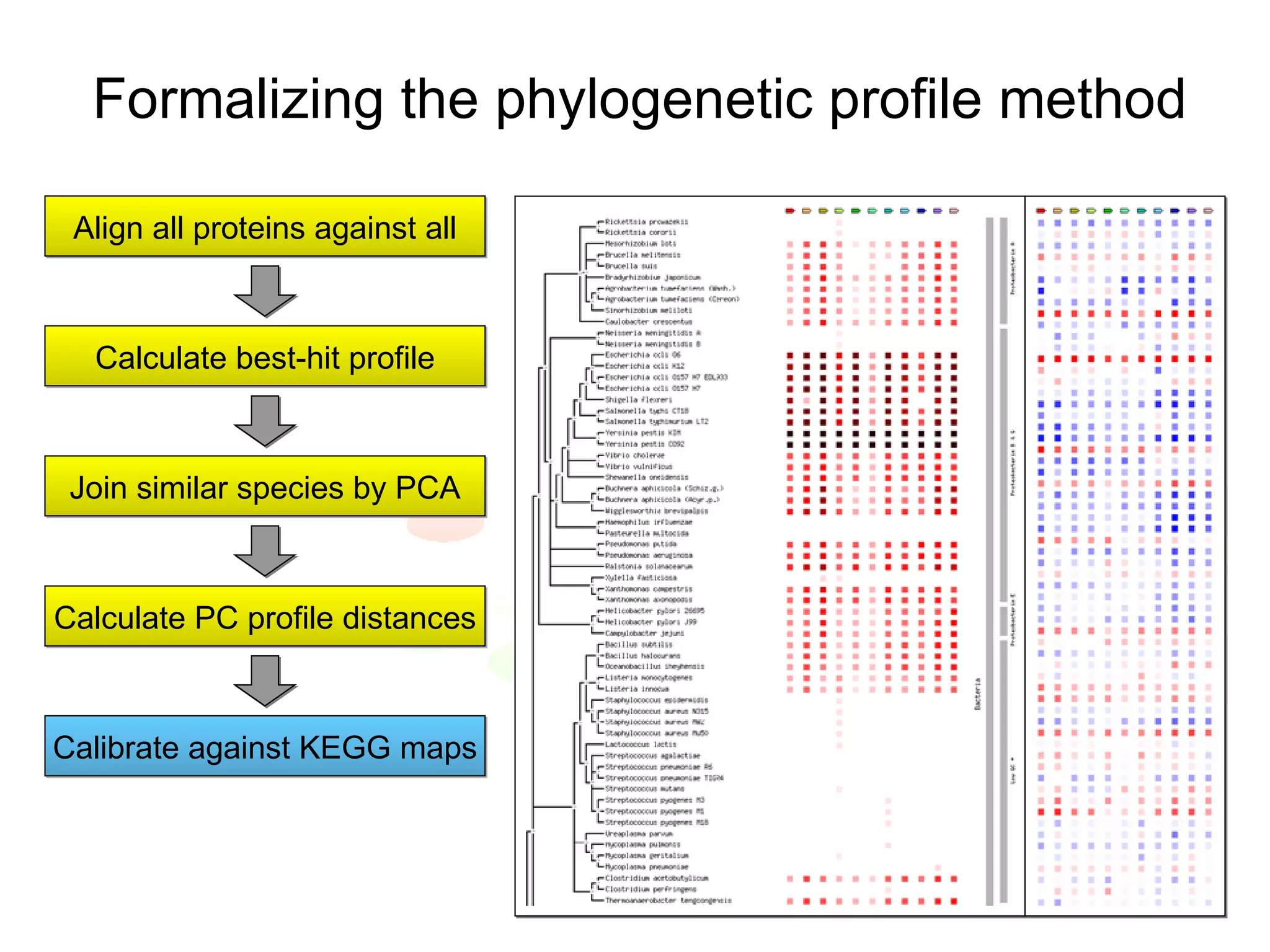
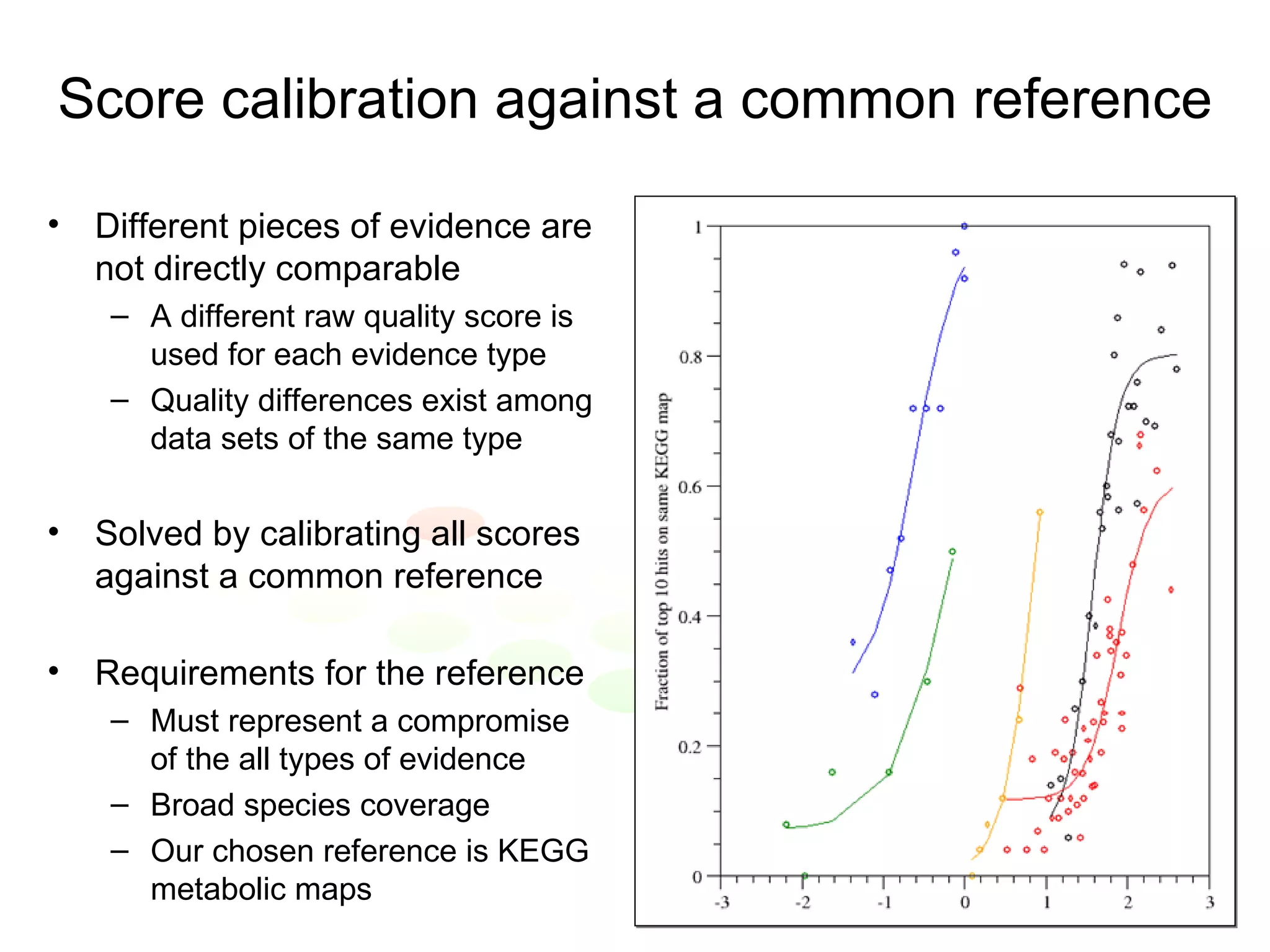
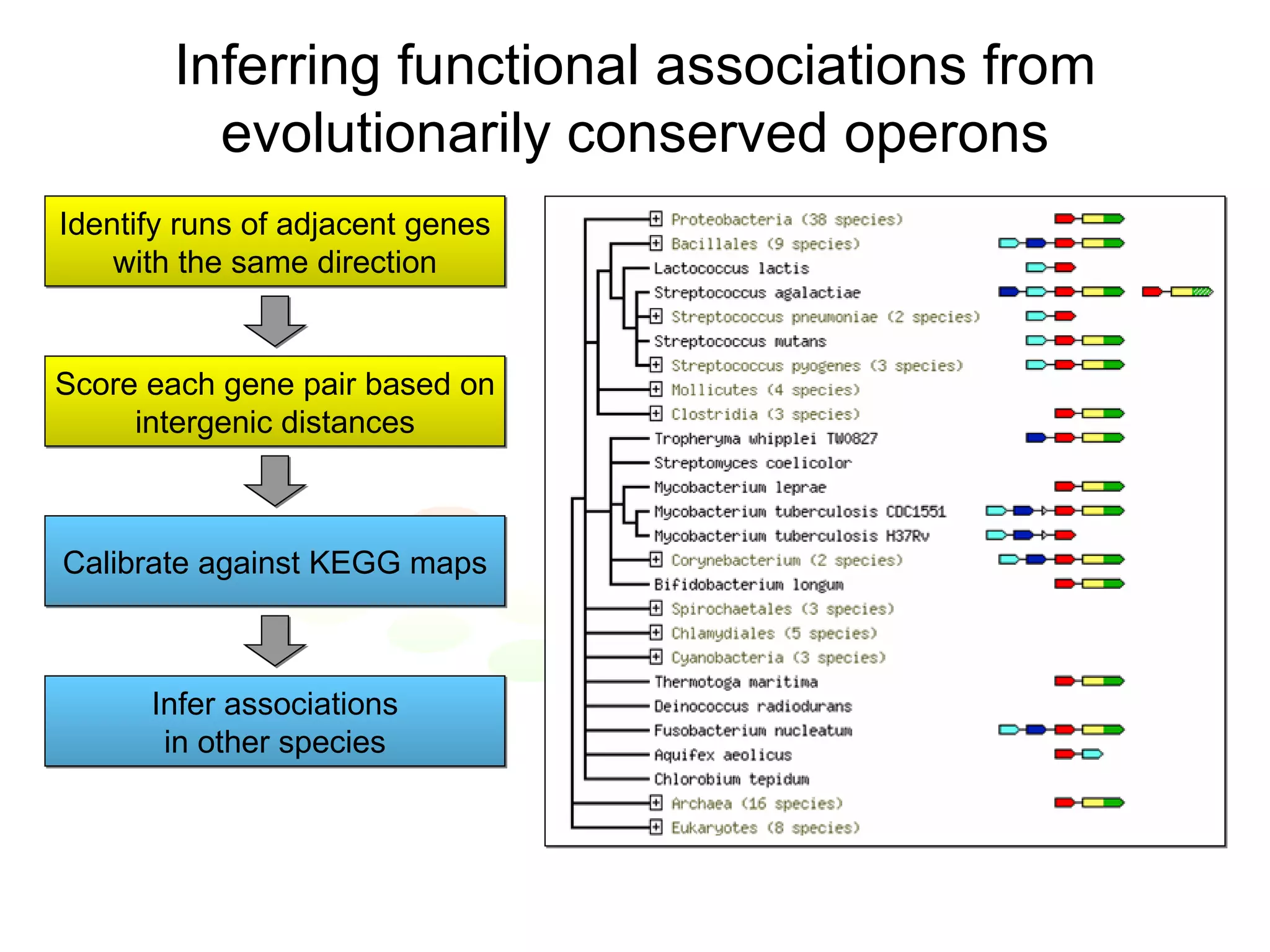
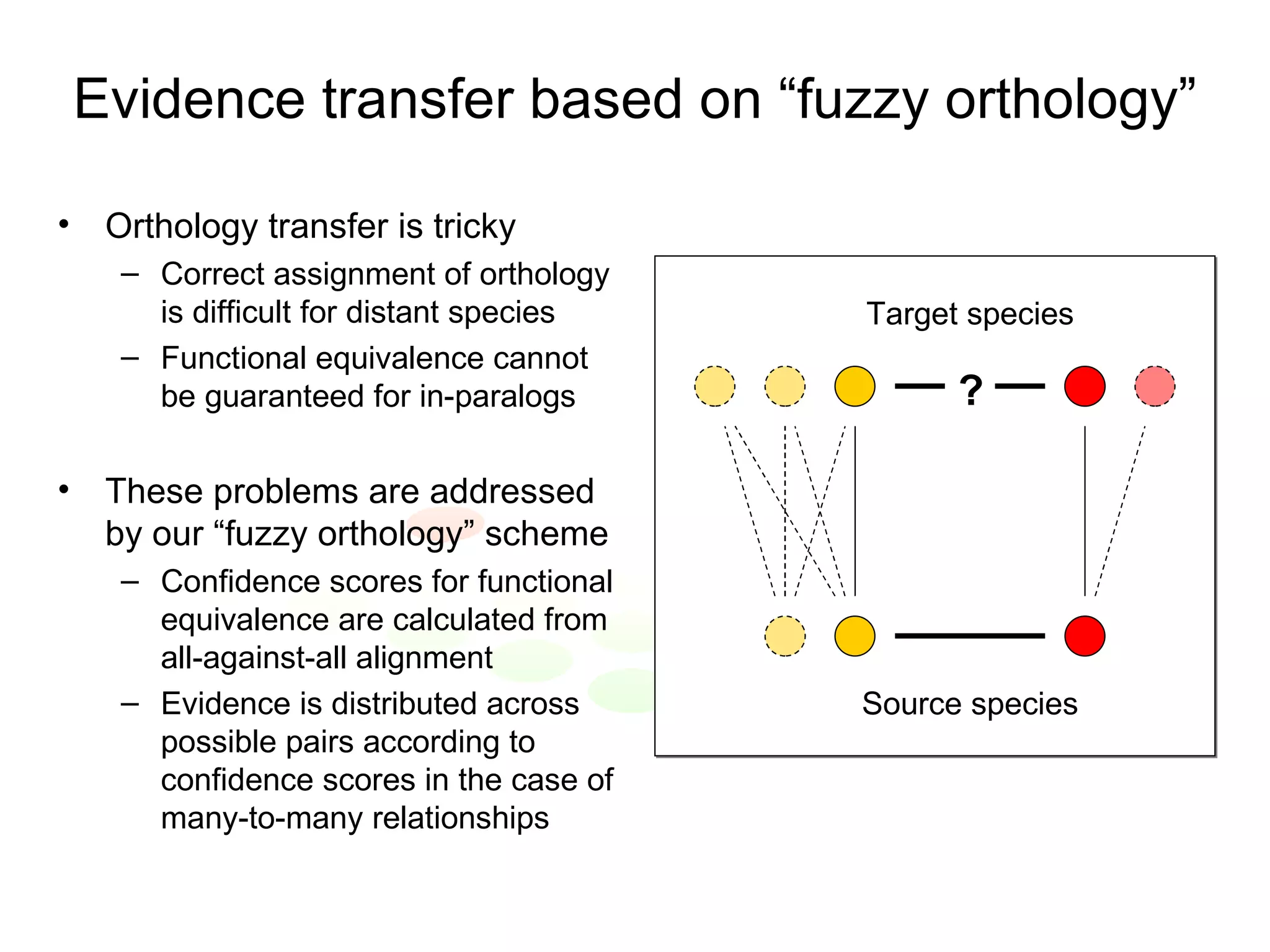
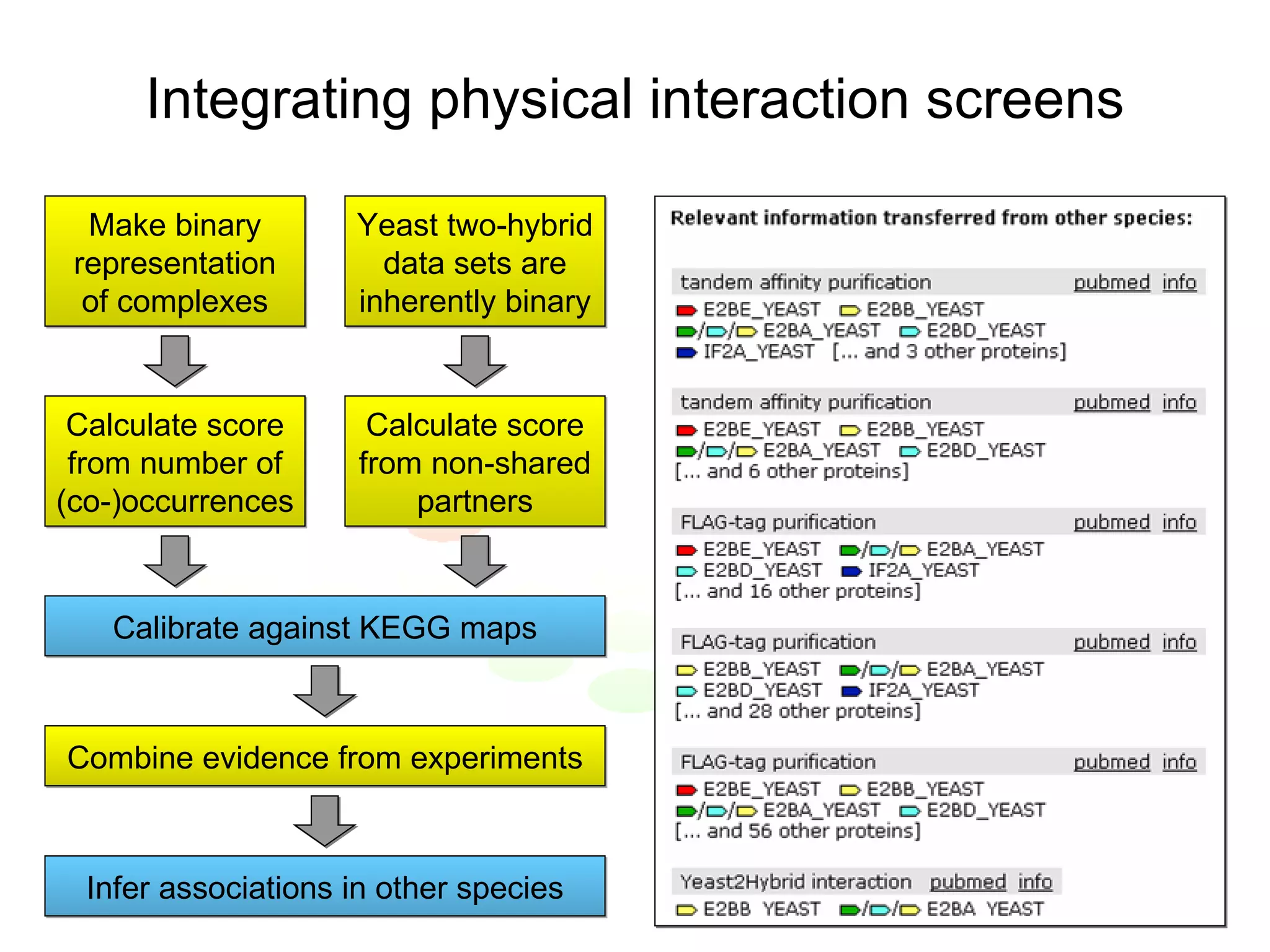
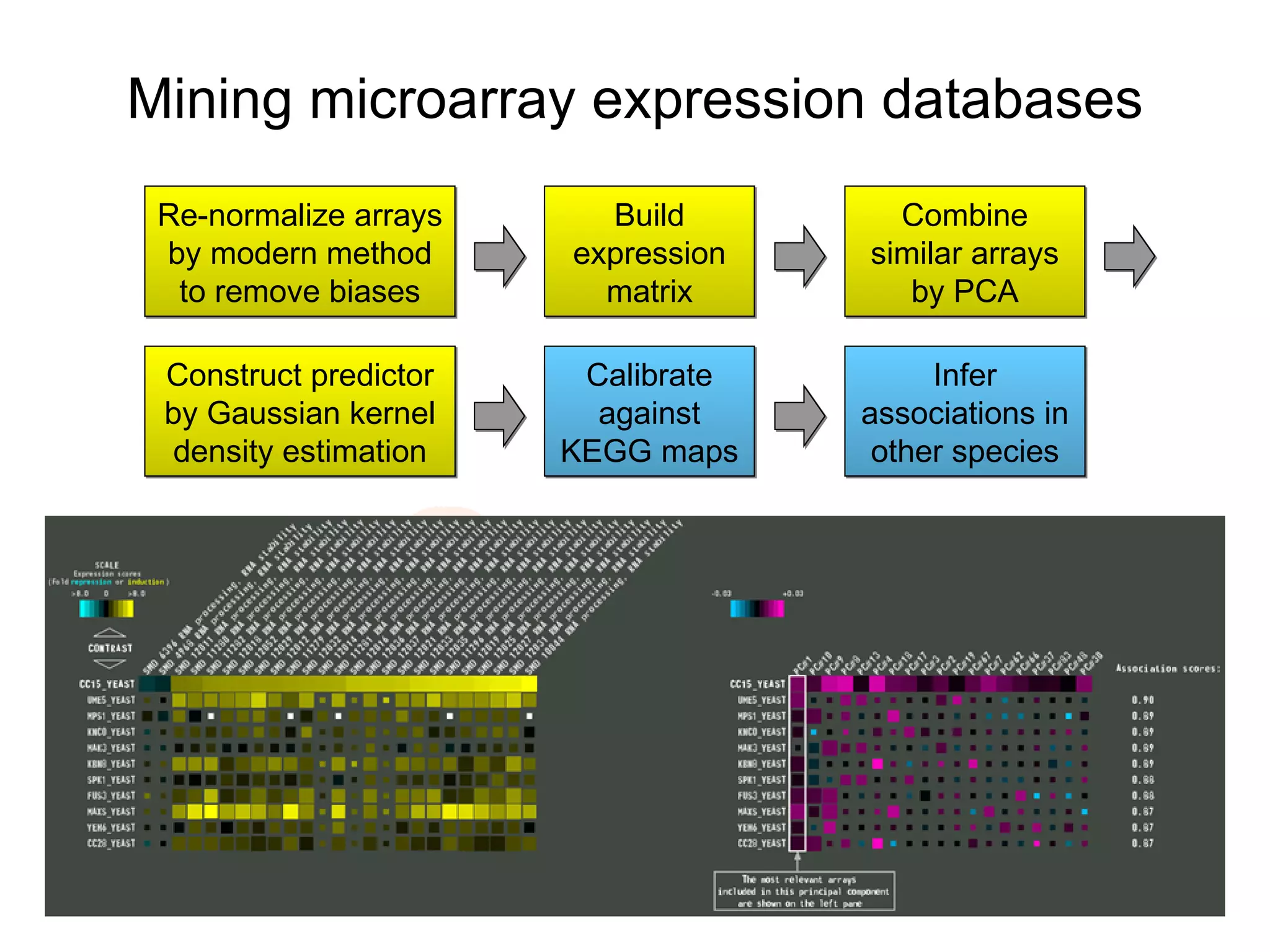
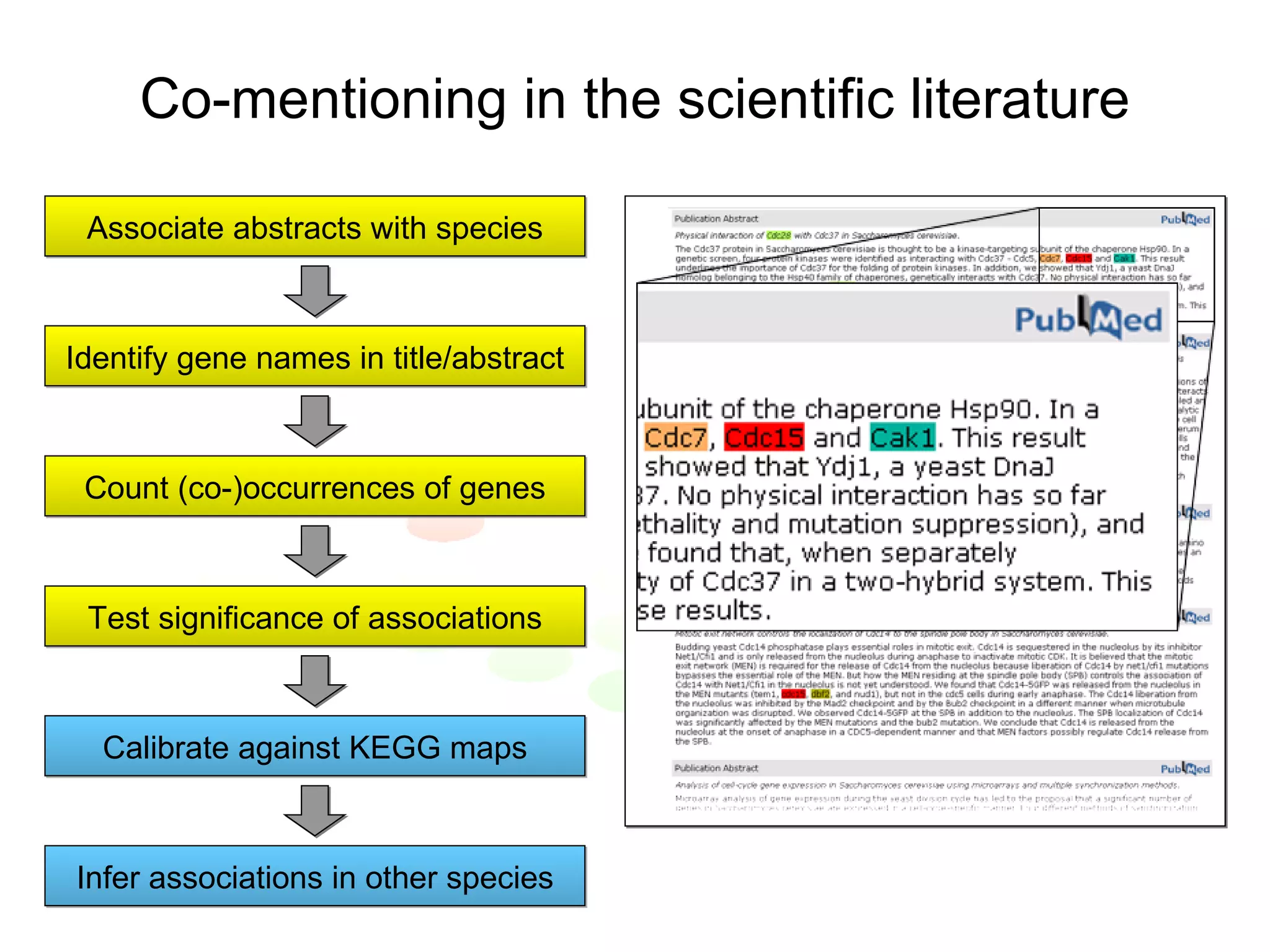
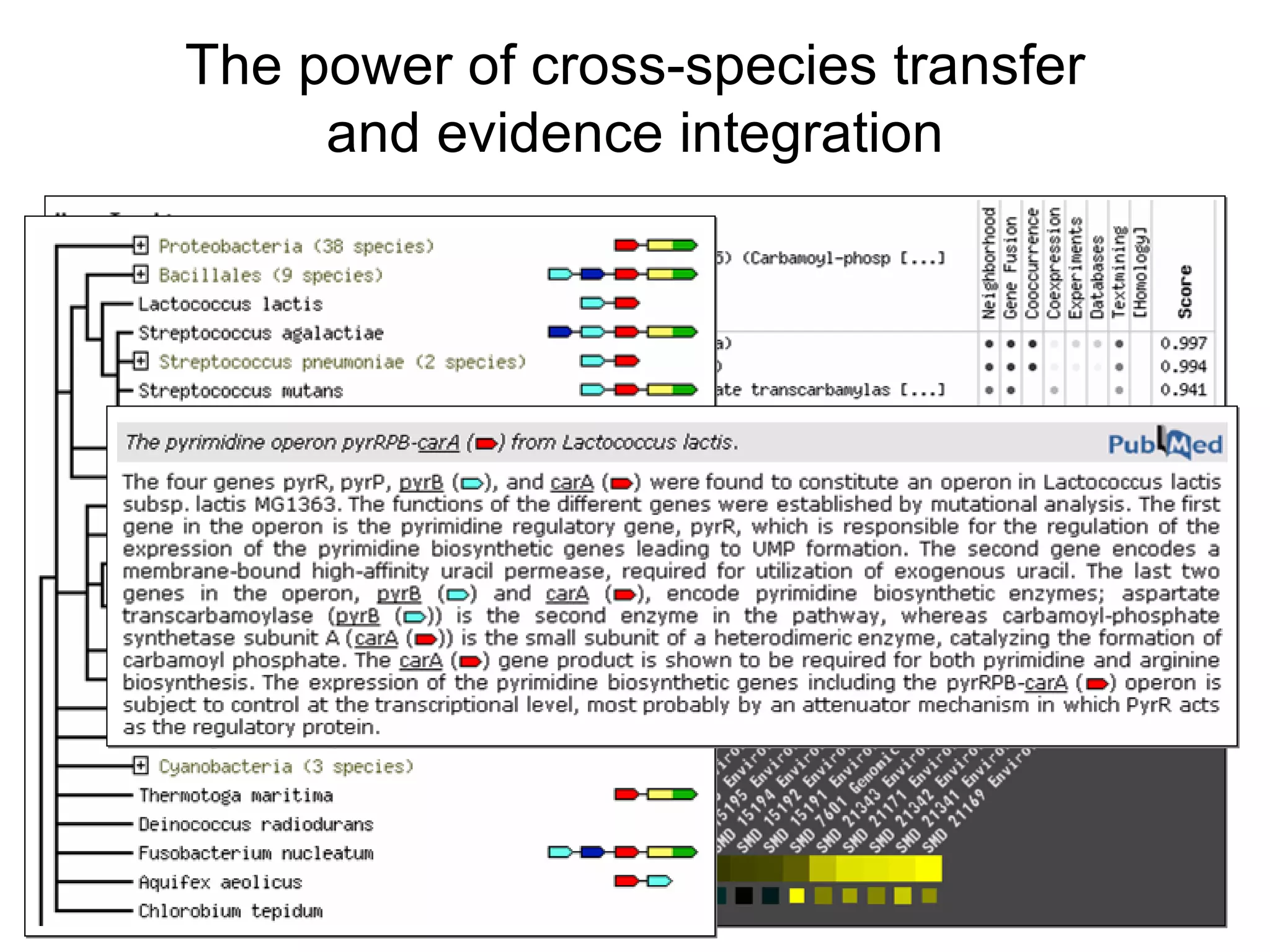
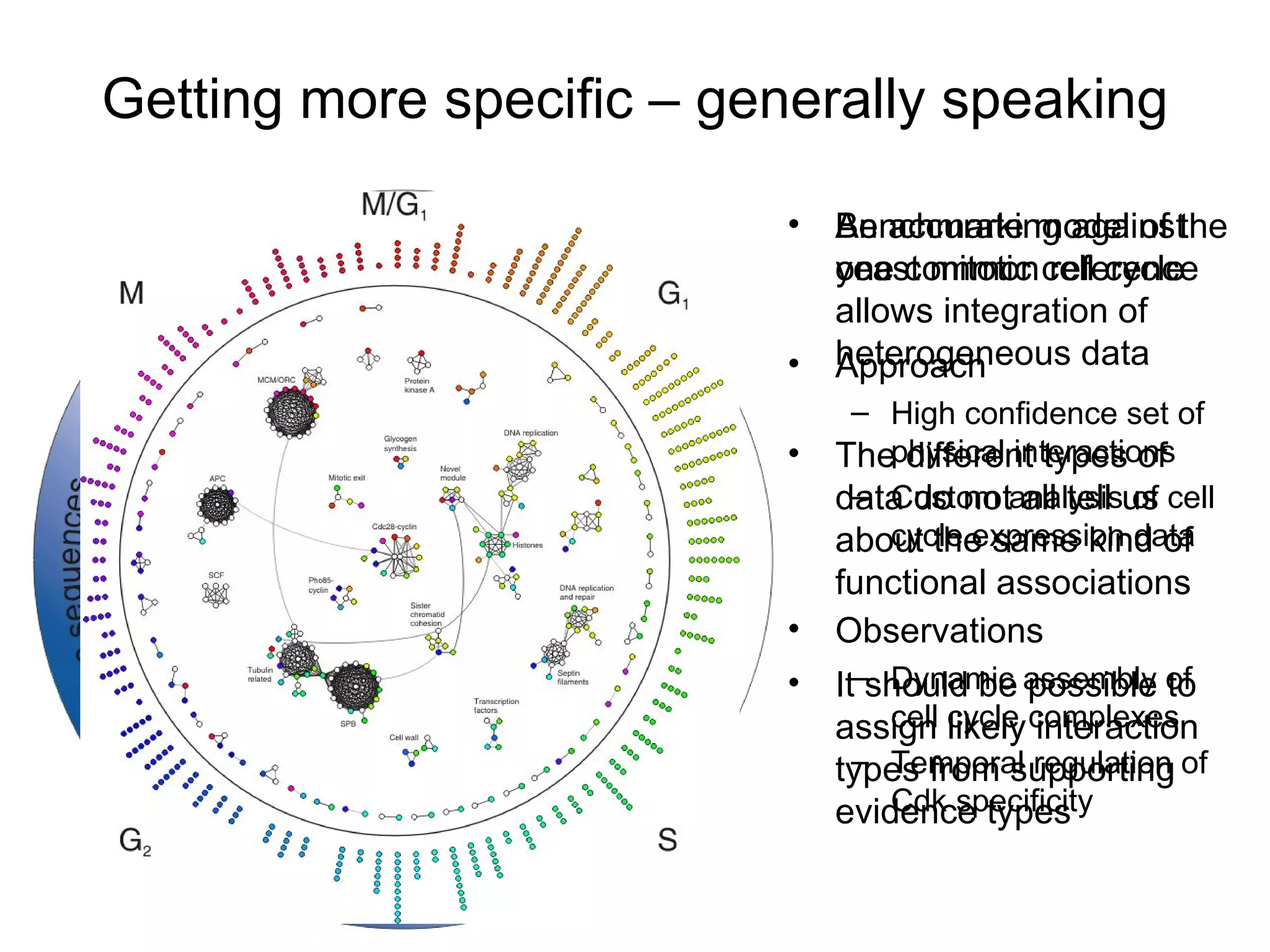
The document discusses the STRING web resource that integrates diverse large-scale genomic data across over 100 species, highlighting the challenges of data heterogeneity, standardization, and biases. It emphasizes the importance of high-quality data integration for predicting functional associations among proteins and proposes methods for inferring relationships through various evidence types. The update to the STRING database aims to enhance cross-species evidence transfer and improve the accuracy of data interpretation.