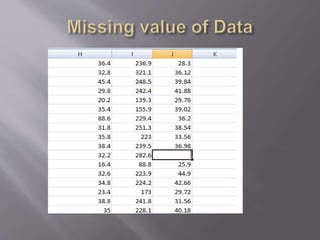
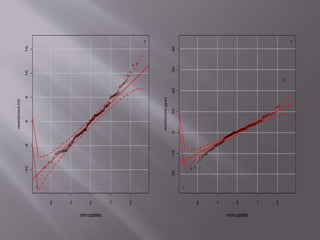
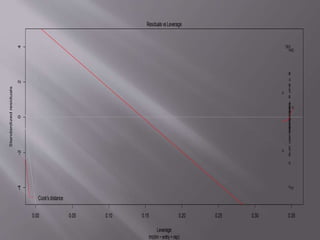
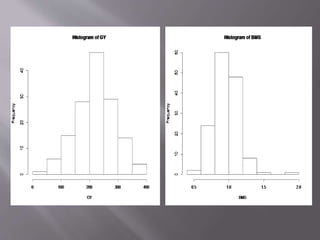
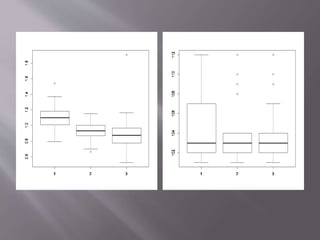
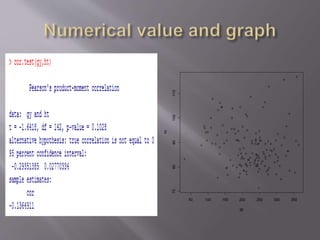
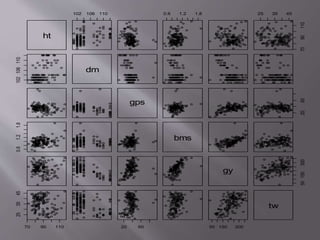
This document provides instructions for using R software for agricultural data analysis. It discusses downloading and installing R, loading and exploring data, performing common analyses like ANOVA and correlations, and using specialized packages for tasks like AMMI analysis. The document recommends exploring R's capabilities and using reliable packages suited for agricultural research.