The document covers parallel computing using R, emphasizing its necessity due to R's default limitation in utilizing multiple cores. It details methodologies for implementing parallel computing through libraries like 'parallel' and 'snow', along with examples like cross-validation and cluster creation. Additional resources and code availability are provided at the end of the document.





![Parallel Computing with R
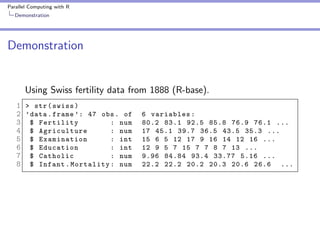
Demonstration
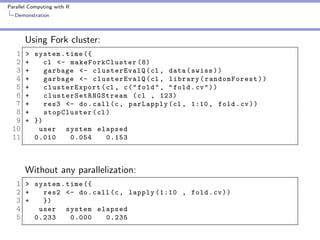
Demonstration
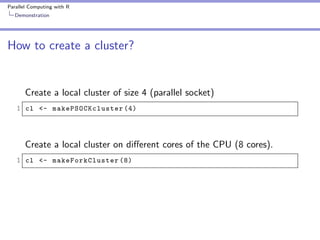
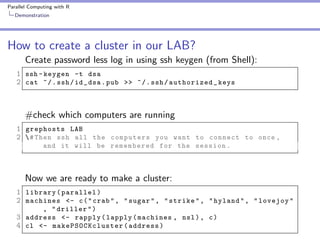
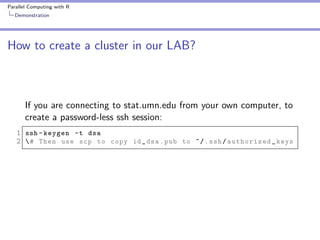
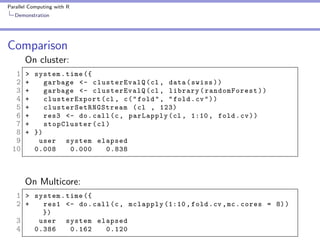
10 fold cross validation
1 fold <- sample ( seq (1 , 10) , size = nrow ( swiss ) ,
2
replace = TRUE )
Cross validation for ’i’th Fold
1 fold . cv <- function ( i ) {
2 train <- swiss [ fold ! = i , ]
3 test <- swiss [ fold == i , ]
4 swiss . rf <- randomForest ( sqrt ( Fertility ) ~ .
5
- Catholic + I ( Catholic < 50) , data = train )
6 predict . test <- predict ( swiss . rf , test , type = " response " )
7 actual . test <- sqrt ( test $ Fertility )
8 err <- predict . test - actual . test
9 sum ( err * err )
10 }](https://image.slidesharecdn.com/presentation-131112204105-phpapp02/85/Parallel-Computing-with-R-12-320.jpg)