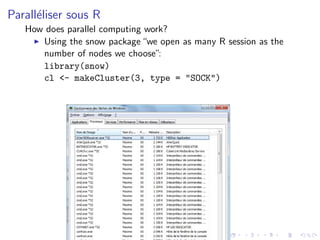
This presentation discusses parallelizing computations in R using the snow package. It demonstrates how to:
1. Create a cluster with multiple R sessions using makeCluster()
2. Split data across the sessions using clusterSplit() and export data to each node
3. Write functions to execute in parallel on each node using clusterEvalQ()
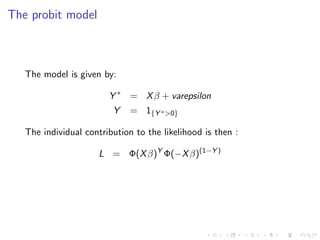
4. Collect the results, such as by summing outputs, to obtain the final parallelized computation. As an example, it shows how to parallelize the likelihood calculation for a probit regression model, reducing the computation time.



![Parall´liser sous R
e
The clusterEvalQ() function allows to execute R code on all
sessions:
clusterEvalQ(cl, ls())
> clusterEvalQ(cl, 1 + 1)
[[1]]
[1] 2
[[2]]
[1] 2
[[3]]
[1] 2](https://image.slidesharecdn.com/pres-120614095529-phpapp01/85/Parallel-R-in-snow-english-after-2nd-slide-5-320.jpg)
![Parall´liser sous R
e
Nodes may be called independently:
> clusterEvalQ(cl[1], a <- 1)
> clusterEvalQ(cl[2], a <- 2)
> clusterEvalQ(cl[3], a <- 3)
> clusterEvalQ(cl, a)
[[1]]
[1] 1
[[2]]
[1] 2
[[3]]
[1] 3](https://image.slidesharecdn.com/pres-120614095529-phpapp01/85/Parallel-R-in-snow-english-after-2nd-slide-6-320.jpg)


![Parall´liser sous R
e
One solution is to first export data to all nodes and then
execute the code on each node:
> #### First Export:
> columns <- clusterSplit(cl, 1:10000)
> for (cc in 1:3){
+ aa <- a[columns[[cc]],]
+ clusterExport(cl[cc], "aa")
+ }
> #### Then execute
>
> system.time(do.call("c",
clusterEvalQ(cl, apply(aa, 1, sum))))
utilisateur syst`me
e e
´coul´
e
0.00 0.00 0.16](https://image.slidesharecdn.com/pres-120614095529-phpapp01/85/Parallel-R-in-snow-english-after-2nd-slide-9-320.jpg)


![A very simple problem
> n <- 5000000
> param <- c(1,2,-.5)
> X1 <- rnorm(n)
> X2 <- rnorm(n, mean = 1, sd = 2)
> Ys <- param[1] + param[2] * X1 +
+ param[3] * X2 + rnorm(n)
> Y <- Ys > 0
> probit <- function(para, y, x1, x2){
+ mu <- para[1] + para[2] * x1 + para[3] * x2
+ sum(pnorm(mu, log = T)*y + pnorm(-mu, log = T)*(1 - y))
+ }
>
> system.time(test1 <- probit(param, Y, X1, X2))
utilisateur syst`me
e e
´coul´
e
1.72 0.08 1.80](https://image.slidesharecdn.com/pres-120614095529-phpapp01/85/Parallel-R-in-snow-english-after-2nd-slide-13-320.jpg)

![Divide data:
> nn <- clusterSplit(cl, 1:n)
> for (cc in 1:3){
+ YY <- Y[nn[[cc]]]
+ XX1 <- X1[nn[[cc]]]
+ XX2 <- X2[nn[[cc]]]
+ clusterExport(cl[cc], c("YY", "XX1", "XX2"))
+ }
> clusterExport(cl, "probit")
> clusterEvalQ(cl, ls())
[[1]]
[1] "probit" "XX1" "XX2" "YY"
[[2]]
[1] "probit" "XX1" "XX2" "YY"
[[3]]
[1] "probit" "XX1" "XX2" "YY"](https://image.slidesharecdn.com/pres-120614095529-phpapp01/85/Parallel-R-in-snow-english-after-2nd-slide-15-320.jpg)

![Execute and compare theg results:
> system.time(test2 <- lik(param)) ## 1.5 sec
utilisateur syst`me
e e
´coul´
e
0.00 0.00 0.78
> c(test1, test2) ## Same results
[1] -1432674 -1432674](https://image.slidesharecdn.com/pres-120614095529-phpapp01/85/Parallel-R-in-snow-english-after-2nd-slide-17-320.jpg)
