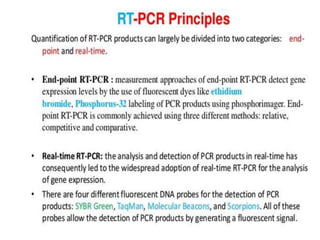
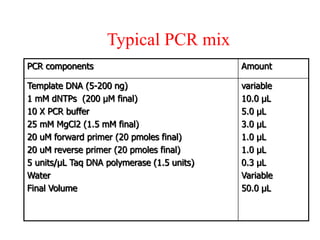
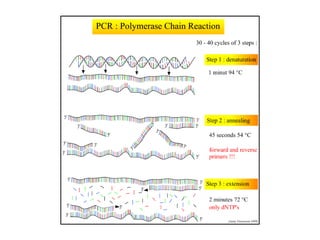
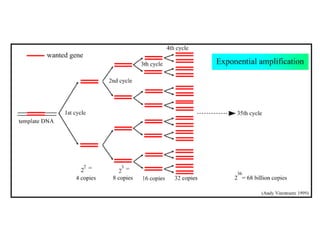
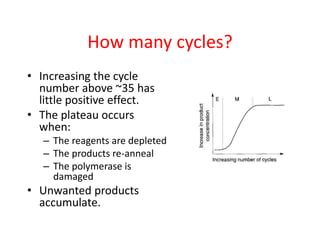
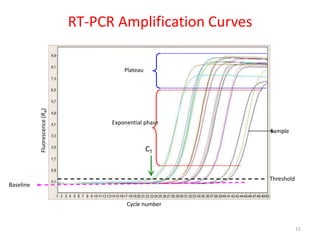
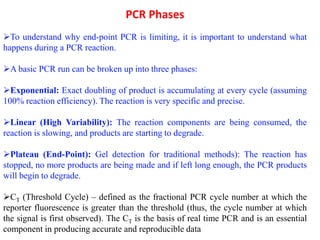
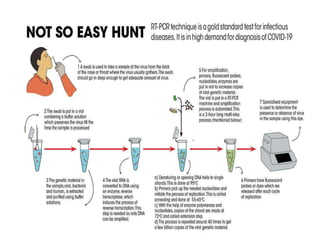
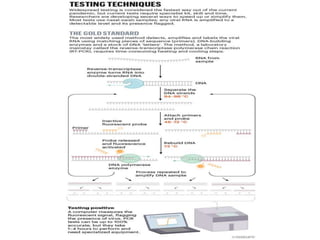
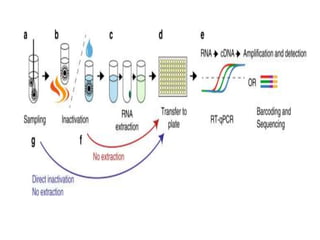
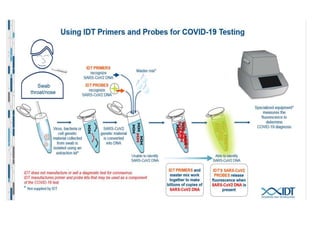
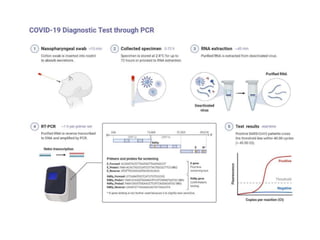
The document discusses polymerase chain reaction (PCR) and its use for detecting COVID-19 through RT-PCR testing. RT-PCR is considered the gold standard for detecting viruses like SARS-CoV-2 due to its rapid detection, high sensitivity, and specificity. The document outlines the RT-PCR testing process, which involves collecting a sample, extracting RNA from the sample, converting the RNA to cDNA, and amplifying the cDNA using fluorescent dyes to detect the presence of the virus. Real-time PCR allows visualization and quantification of PCR product accumulation in real-time by using fluorescent dyes that bind to DNA.