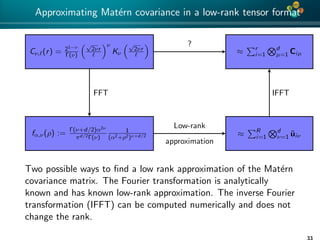
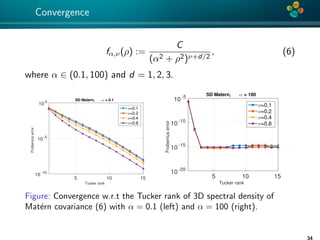
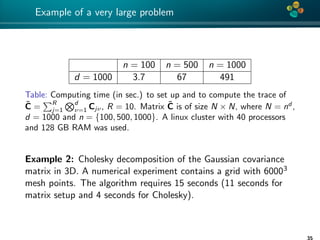
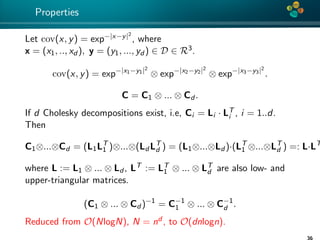
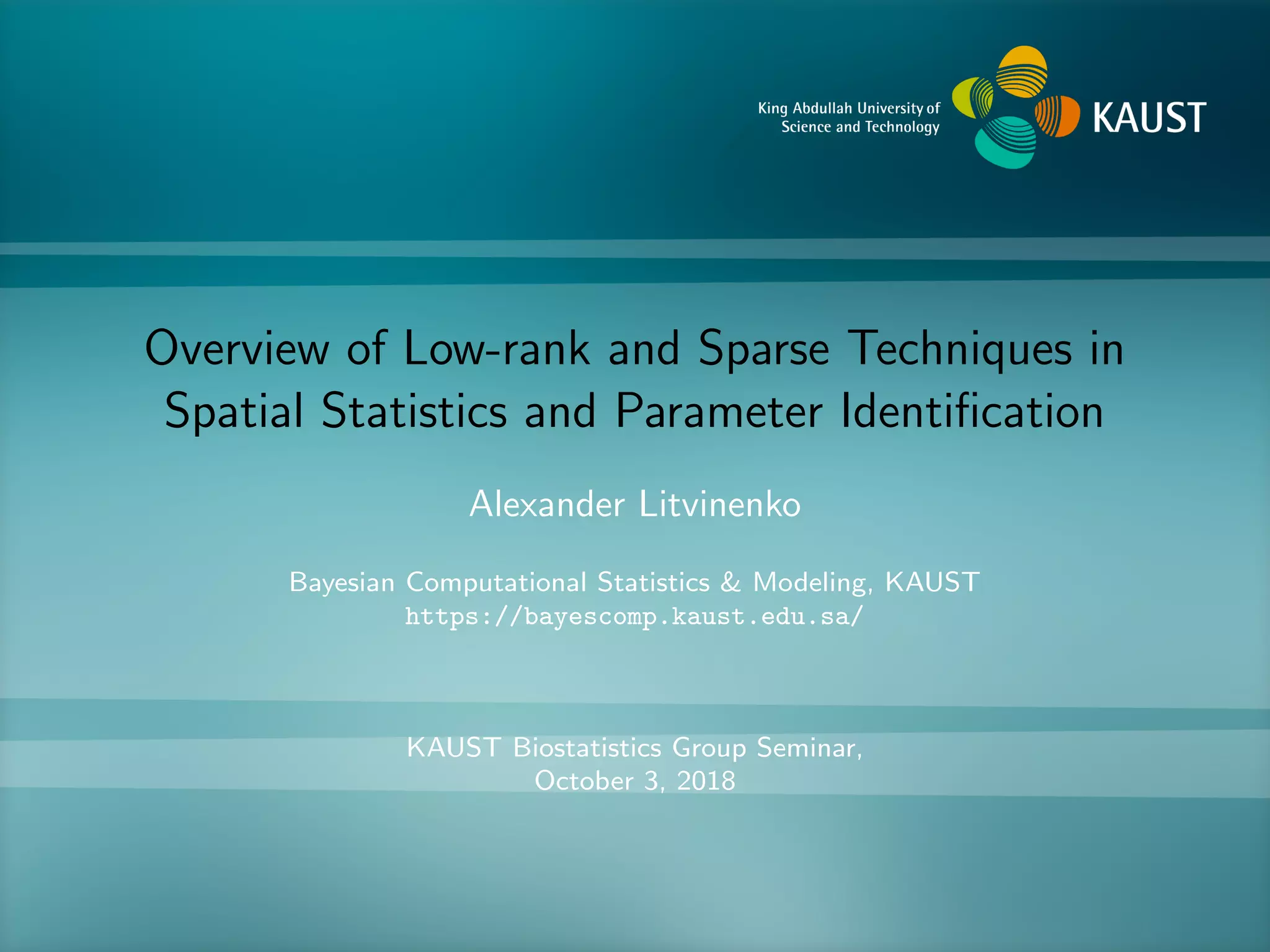
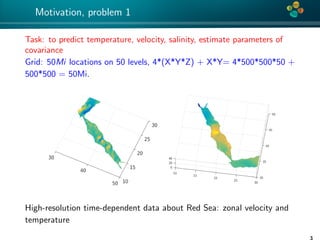
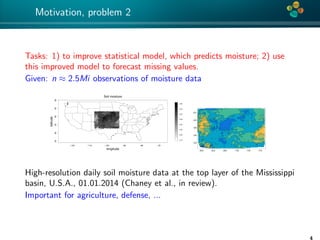
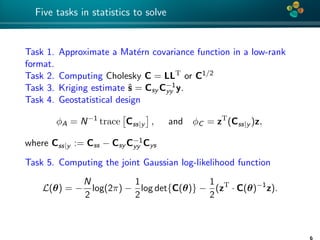
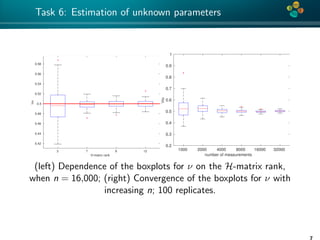
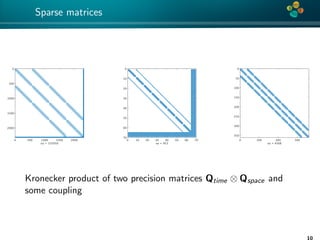
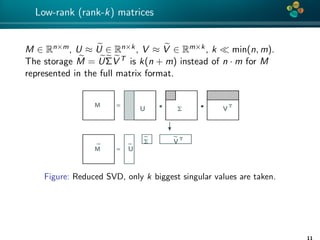
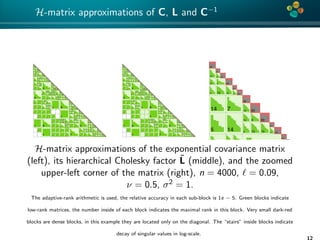
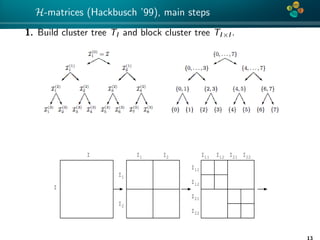
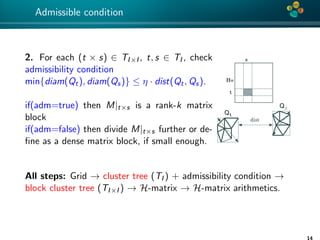
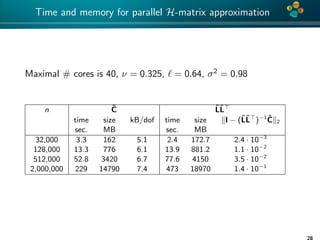
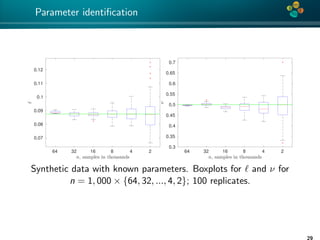
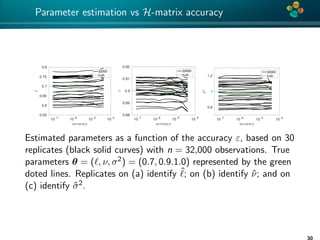
The document presents an overview of low-rank and sparse techniques in spatial statistics and parameter identification, highlighting their applications in predicting environmental variables. It addresses key problems such as predicting temperature and salinity, improving statistical models for moisture, and estimating covariance parameters. The document also outlines various statistical tasks and the techniques available, including sparse matrices and low-rank methods.


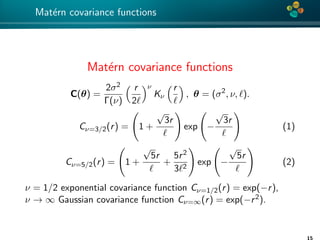

![4*
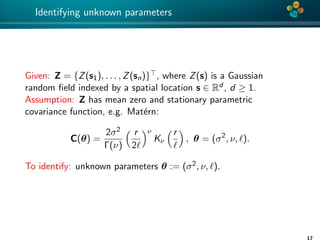
Details of the identification
To maximize the log-likelihood function we use the Brent’s method
[Brent’73] (combining bisection method, secant method and
inverse quadratic interpolation).
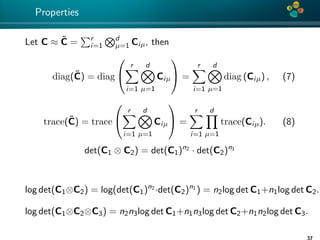
1. H-matrix: C(θ) ≈ C(θ, k) or ≈ C(θ, ε).
2. H-Cholesky: C(θ, k) = L(θ, k)L(θ, k)T
3. ZT C−1Z = ZT (LLT )−1Z = vT · v, where v is a solution of
L(θ, k)v(θ) := Z.
log det{C} = log det{LLT
} = log det{
n
i=1
λ2
i } = 2
n
i=1
logλi ,
L(θ, k) = −
N
2
log(2π) −
N
i=1
log{Lii (θ, k)} −
1
2
(v(θ)T
· v(θ)). (4)
19](https://image.slidesharecdn.com/talklitvinenkoombao-181004065628/85/Overview-of-sparse-and-low-rank-matrix-tensor-techniques-19-320.jpg)
![4*
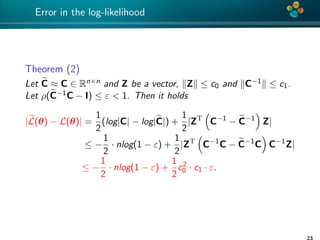
Error analysis
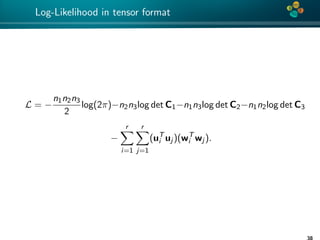
Theorem (1)
Let C be an H-matrix approximation of matrix C ∈ Rn×n such that
ρ(C−1
C − I) ≤ ε < 1.
Then
|log|C| − log|C|| ≤ −nlog(1 − ε), (5)
Proof: See [Ballani, Kressner 14] and [Ipsen 05].
Remark: factor n is pessimistic and is not really observed
numerically.
22](https://image.slidesharecdn.com/talklitvinenkoombao-181004065628/85/Overview-of-sparse-and-low-rank-matrix-tensor-techniques-22-320.jpg)
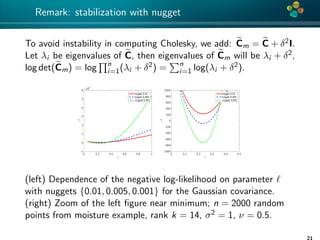
![4*
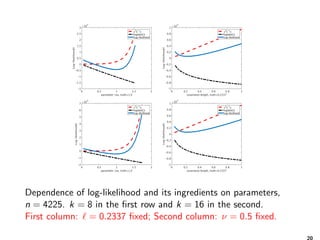
How much memory is needed?
0 0.5 1 1.5 2 2.5
ℓ, ν = 0.325, σ2
= 0.98
5
5.5
6
6.5
size,inbytes
×10 6
1e-4
1e-6
0.2 0.4 0.6 0.8 1 1.2 1.4
ν, ℓ = 0.58, σ2
= 0.98
5.2
5.4
5.6
5.8
6
6.2
6.4
6.6
size,inbytes
×10 6
1e-4
1e-6
(left) Dependence of the matrix size on the covariance length ,
and (right) the smoothness ν for two different accuracies in the
H-matrix sub-blocks ε = {10−4, 10−6}, for n = 2, 000 locations in
the domain [32.4, 43.4] × [−84.8, −72.9].
25](https://image.slidesharecdn.com/talklitvinenkoombao-181004065628/85/Overview-of-sparse-and-low-rank-matrix-tensor-techniques-25-320.jpg)
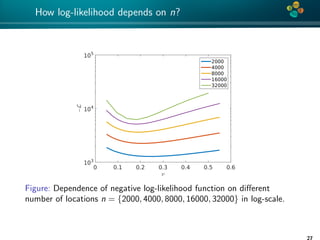
![4*
Error convergence
0 10 20 30 40 50 60 70 80 90 100
−25
−20
−15
−10
−5
0
rank k
log(rel.error)
Spectral norm, L=0.1, nu=1
Frob. norm, L=0.1
Spectral norm, L=0.2
Frob. norm, L=0.2
Spectral norm, L=0.5
Frob. norm, L=0.5
0 10 20 30 40 50 60 70 80 90 100
−16
−14
−12
−10
−8
−6
−4
−2
0
rank k
log(rel.error)
Spectral norm, L=0.1, nu=0.5
Frob. norm, L=0.1
Spectral norm, L=0.2
Frob. norm, L=0.2
Spectral norm, L=0.5
Frob. norm, L=0.5
Convergence of the H-matrix approximation errors for covariance
lengths {0.1, 0.2, 0.5}; (left) ν = 1 and (right) ν = 0.5,
computational domain [0, 1]2.
26](https://image.slidesharecdn.com/talklitvinenkoombao-181004065628/85/Overview-of-sparse-and-low-rank-matrix-tensor-techniques-26-320.jpg)

![4*
Canonical (left) and Tucker (right) decompositions of 3D tensors.
R
(3)
1
(2)
2
(1)
1
(3)
R
(2)
R
(1)
R
.
.
b
c1
u
u
u u
u
u
c+ . . . +
A
n3
n1
n2
r3
3
2
r
1
r
r
r1
B
1
3
2
r
2
A
V
(1)
V
(2)
V
(3)
n
n
n
n
1
n
3
n
2
I
I
I
2
3
1
I3
A
I2
A(1)I
1
[A. Litvinenko, D. Keyes, V. Khoromskaia, B. N. Khoromskij, H. G. Matthies, Tucker Tensor Analysis of Mat´ern
Functions in Spatial Statistics, 2018]
32](https://image.slidesharecdn.com/talklitvinenkoombao-181004065628/85/Overview-of-sparse-and-low-rank-matrix-tensor-techniques-32-320.jpg)