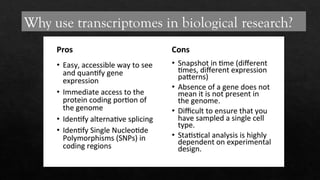
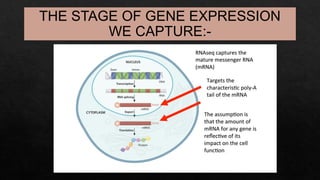
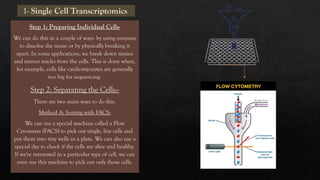
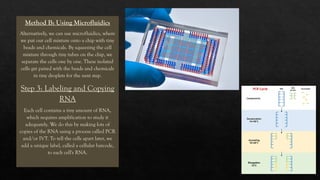
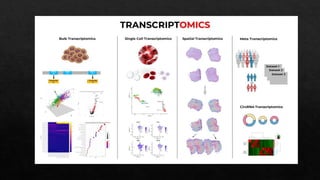
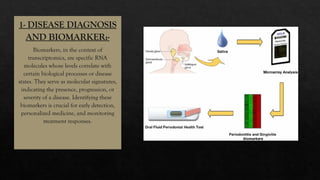
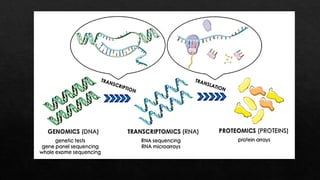
The document discusses transcriptomics, the study of RNA molecules in cells, including various methodologies like single-cell transcriptomics and high-throughput sequencing. It outlines the steps involved in analyzing RNA, from cell preparation and separation to data analysis and visualization, highlighting its applications in disease diagnosis, drug development, cancer research, and plant breeding. The potential for identifying biomarkers and key regulatory genes is emphasized as a critical advancement in biological research.