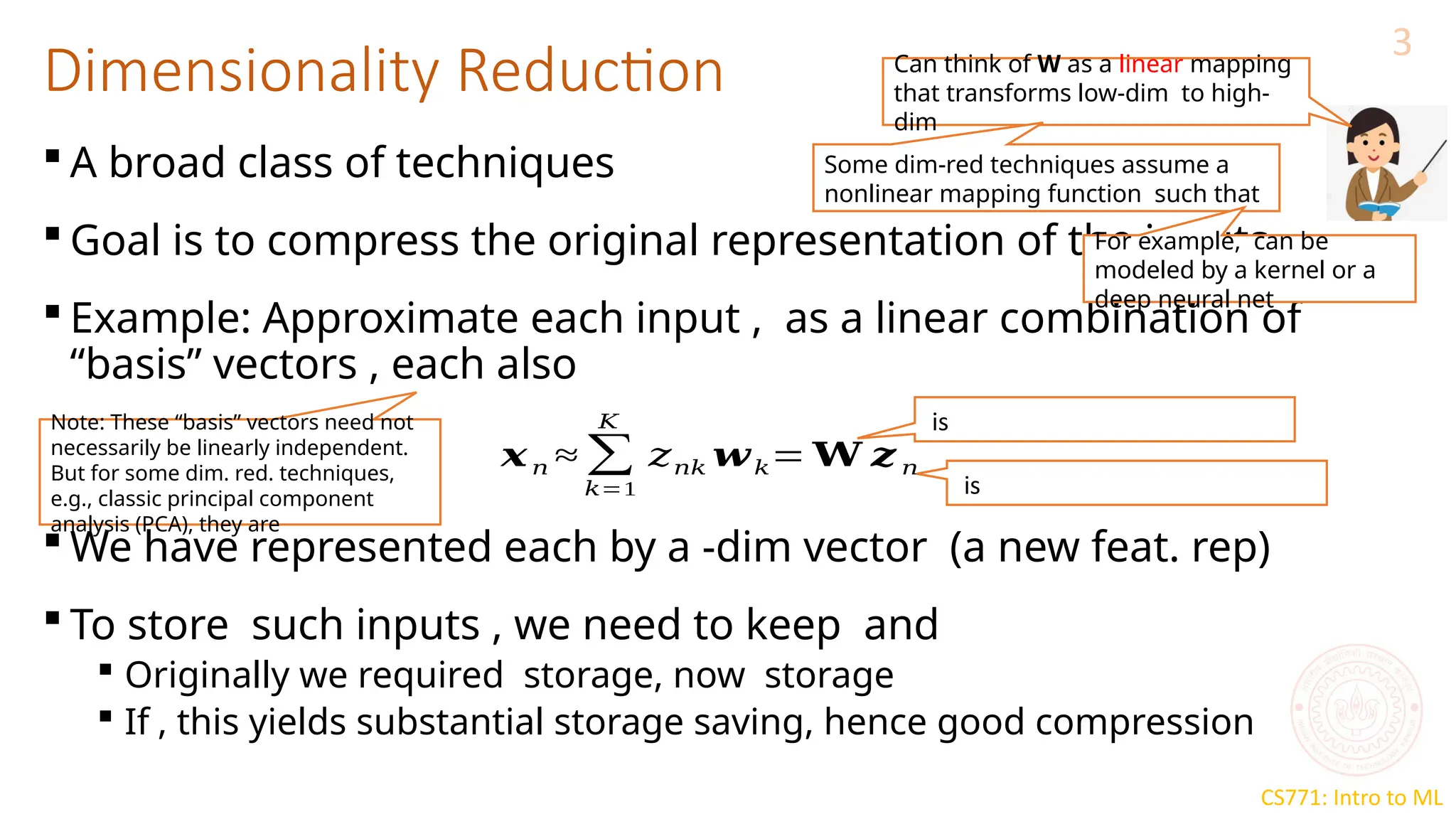
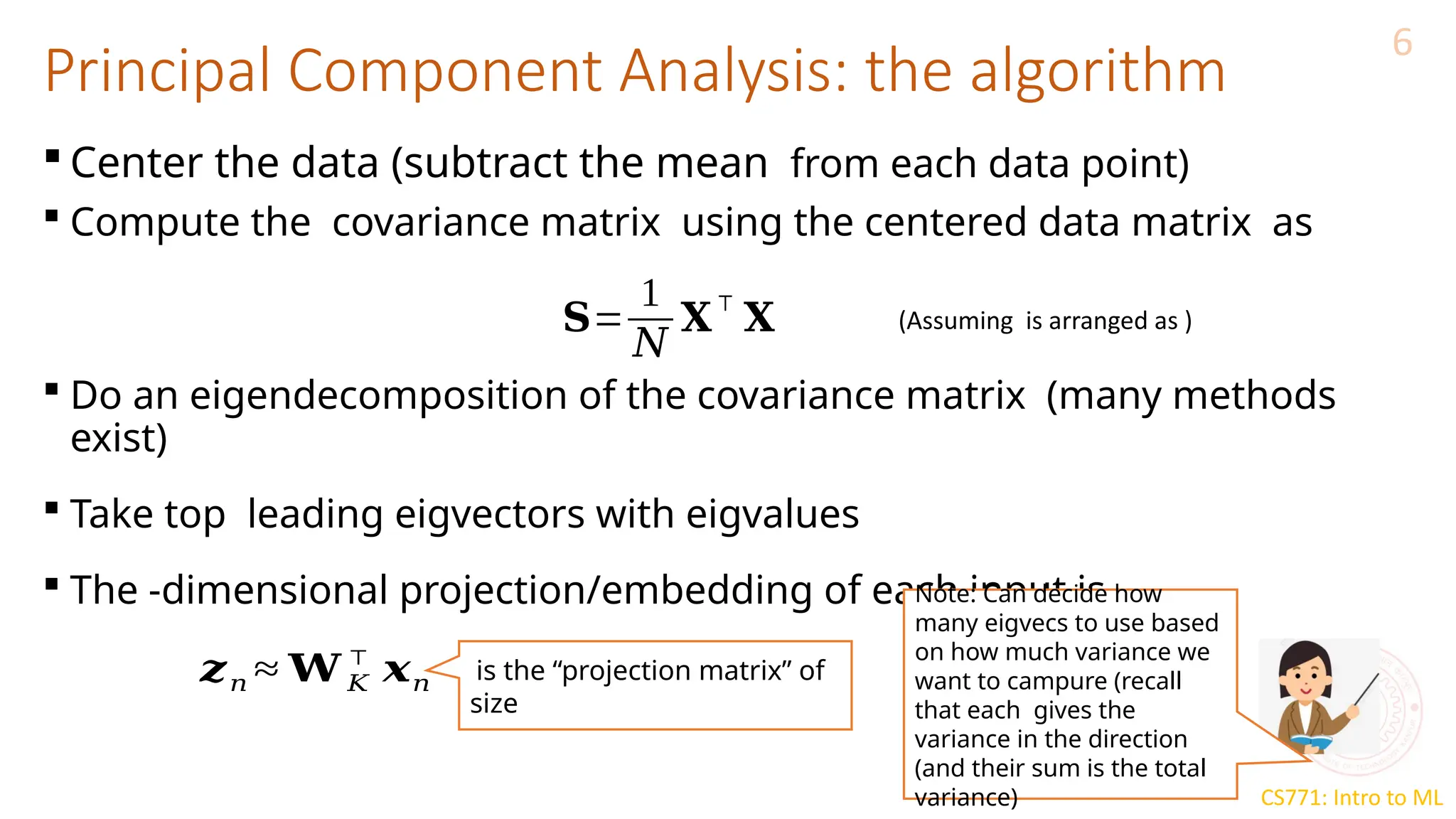
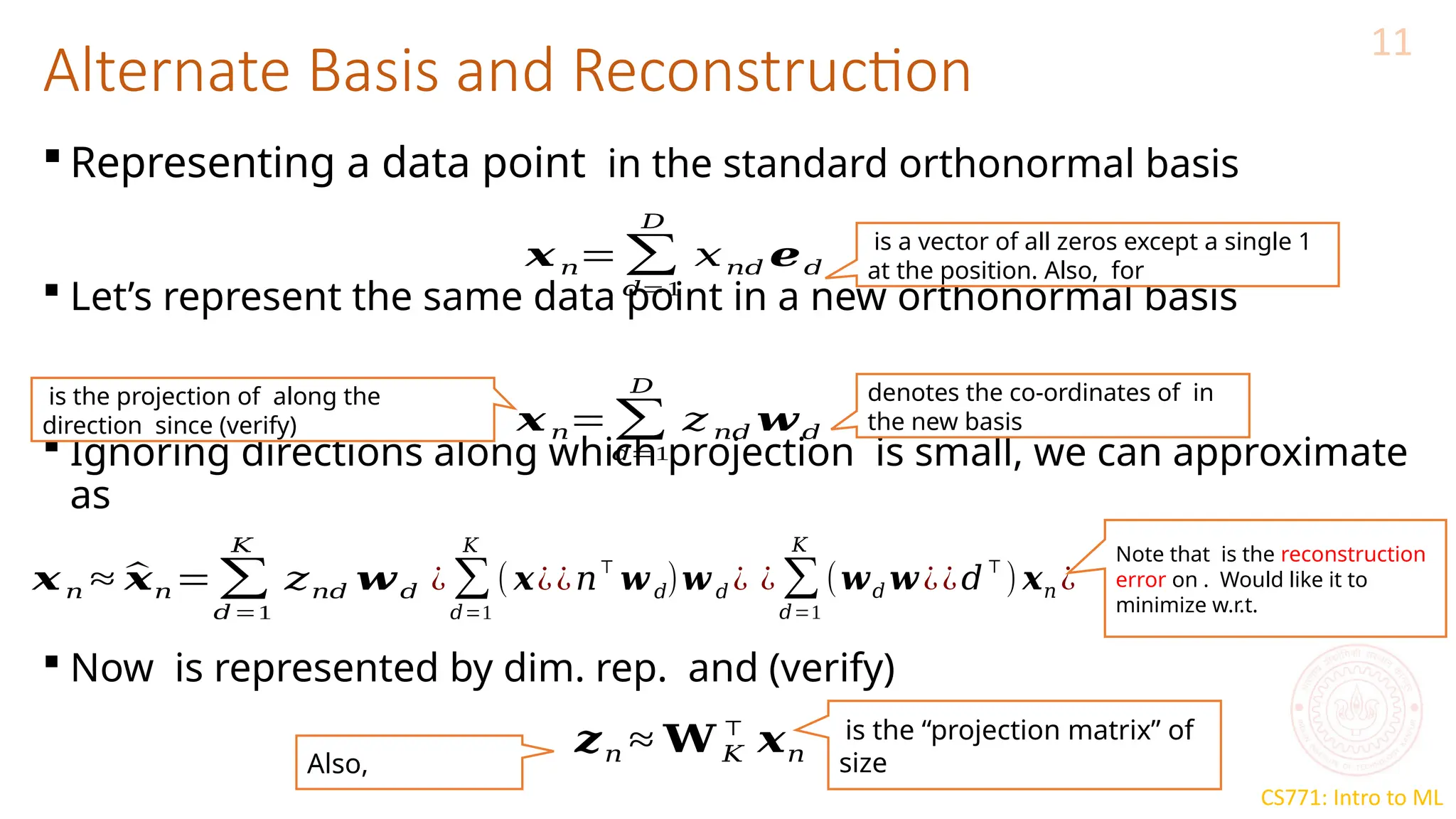
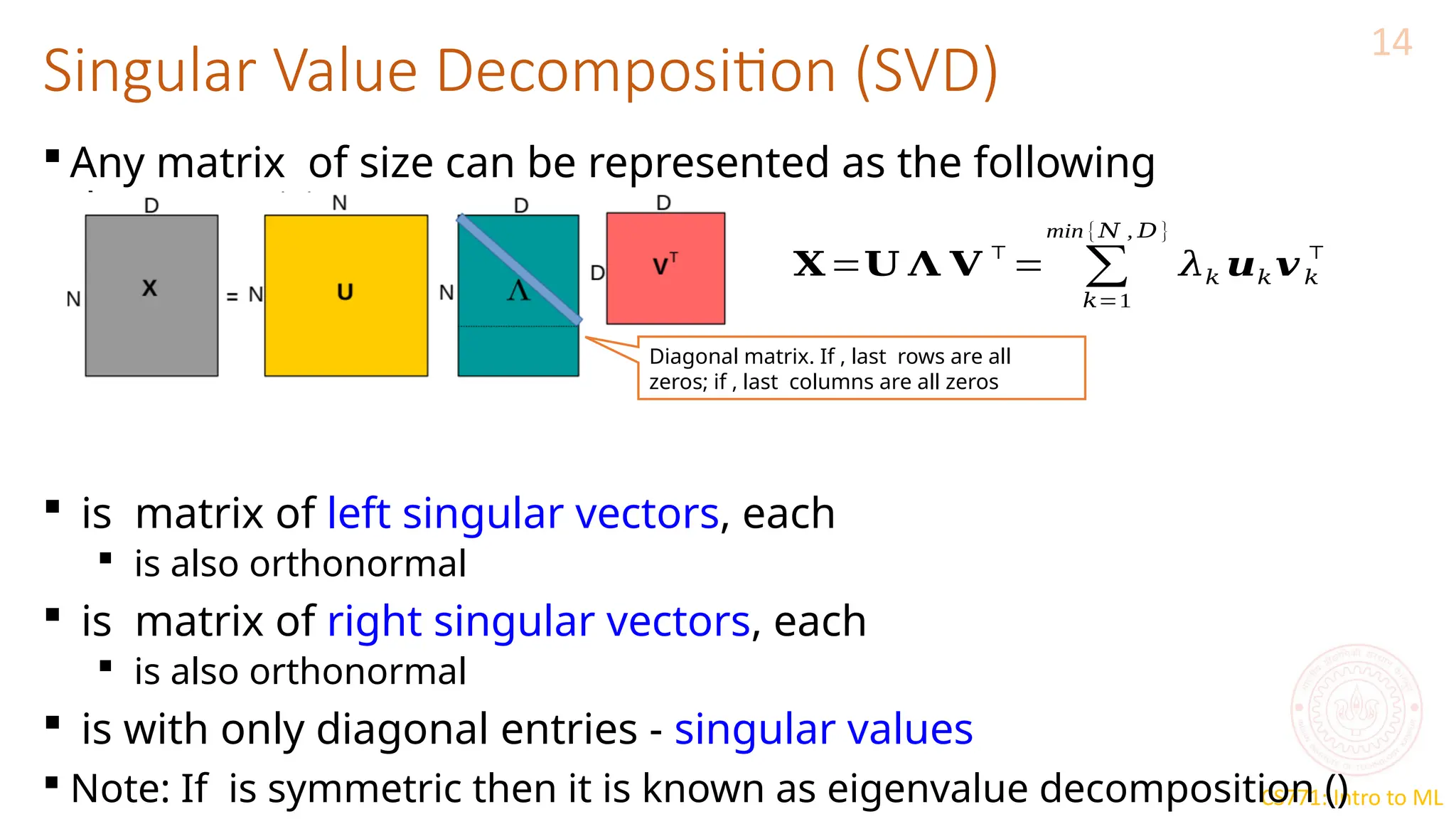
The document discusses dimensionality reduction techniques, particularly Principal Component Analysis (PCA), which aims to compress data while preserving its variance. PCA is introduced as a linear method that identifies directions of maximum variance in the data by performing eigendecomposition on the covariance matrix, allowing for efficient data representation with lower dimensions. Additionally, Singular Value Decomposition (SVD) is mentioned as a method to achieve low-rank approximations of matrices and facilitate dimensionality reduction.

![CS771: Intro to ML
K-means loss function: recap
2
X Z
N
K
K
[, ] denotes a length
one-hot encoding of
• Remember the matrix factorization view of the k-means loss function?
• We approximated an N x D matrix with
• An NxK matrix and a
• KXD matrix
• This could be storage efficient if K is much smaller than D
D
D](https://image.slidesharecdn.com/lec22-241216041716-66c16bba/75/lec22-pca-DIMENSILANITY-REDUCTION-pptx-2-2048.jpg)