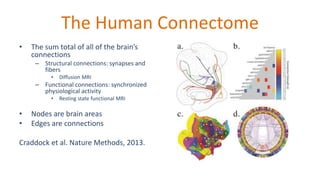
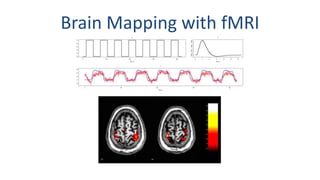
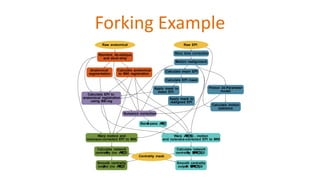
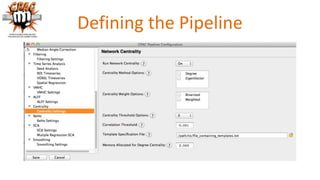
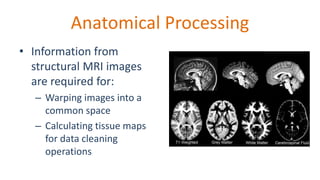
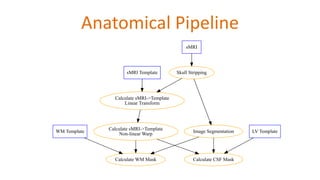
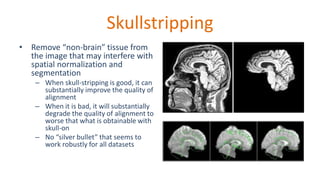
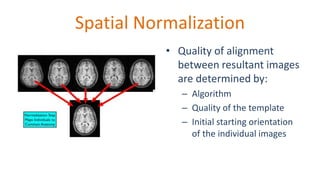
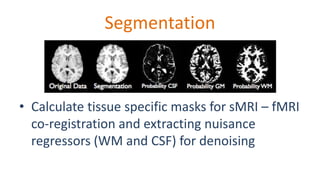
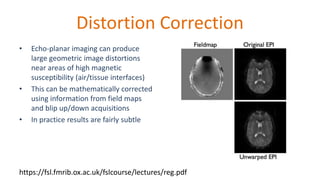
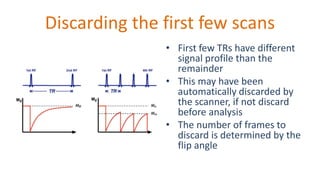
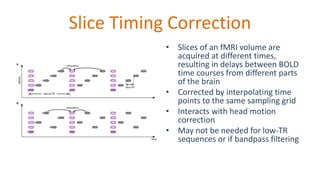
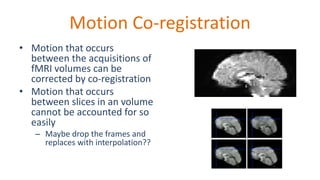
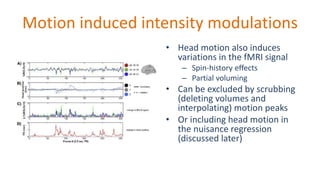
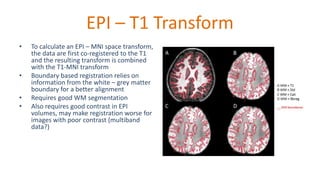
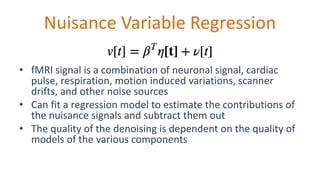
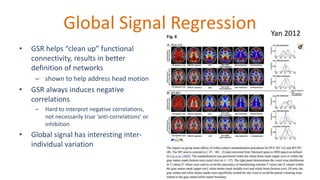
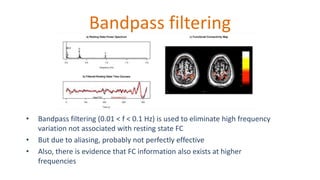
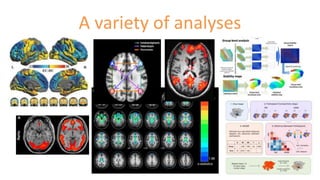
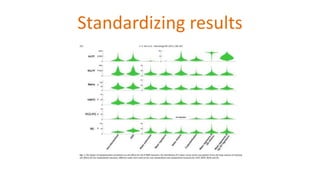
The document outlines the methodologies and best practices for analyzing functional connectomes using resting-state functional MRI (r-fMRI) and diffusion MRI, detailing processes such as preprocessing, nuisance variable regression, and noise correction. It emphasizes the importance of addressing physiological noise and the challenges related to head motion in fMRI studies. Additionally, it discusses various analysis techniques, standardizations, and the integration of structural MRI data for improved spatial normalization.