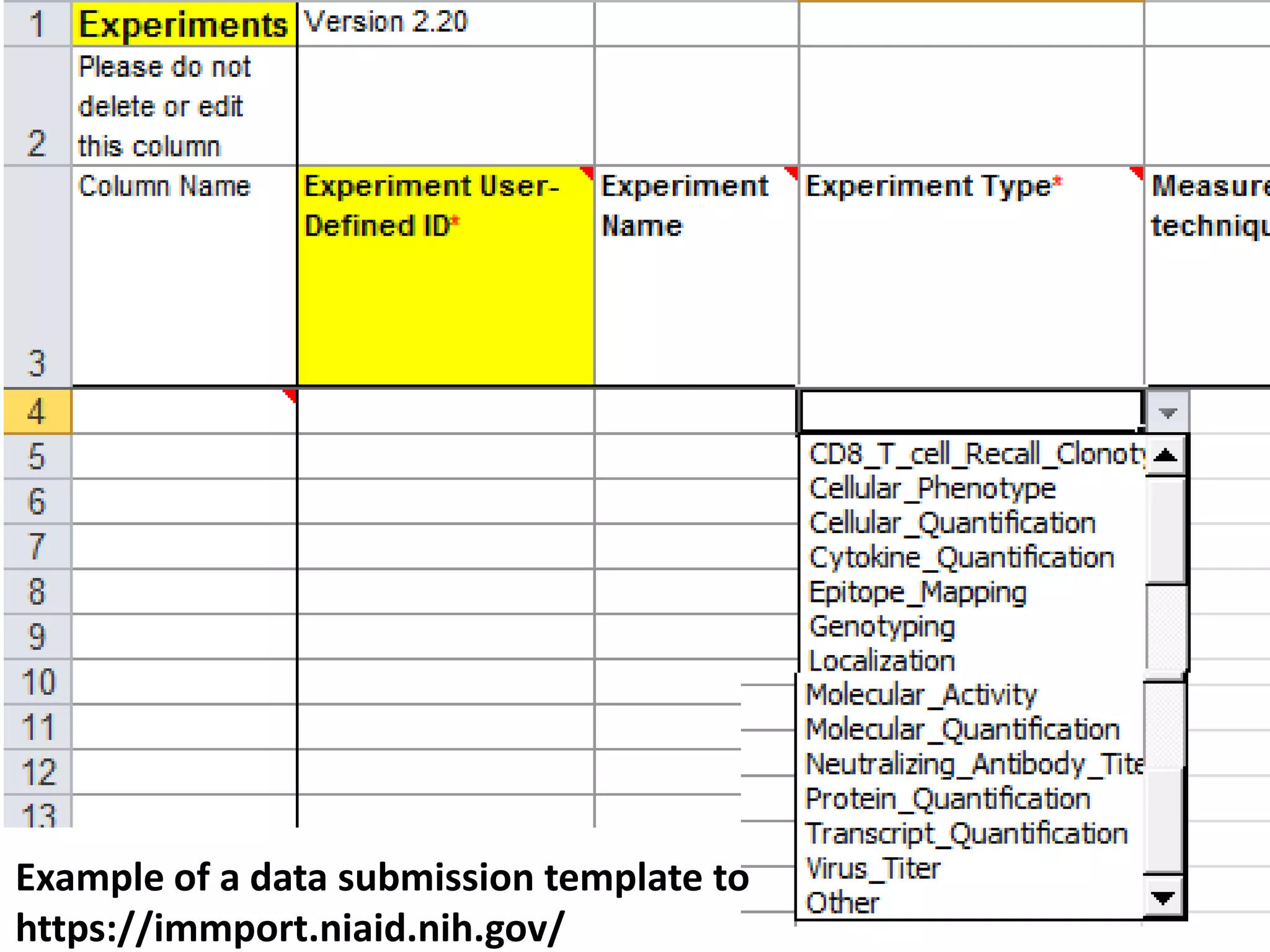
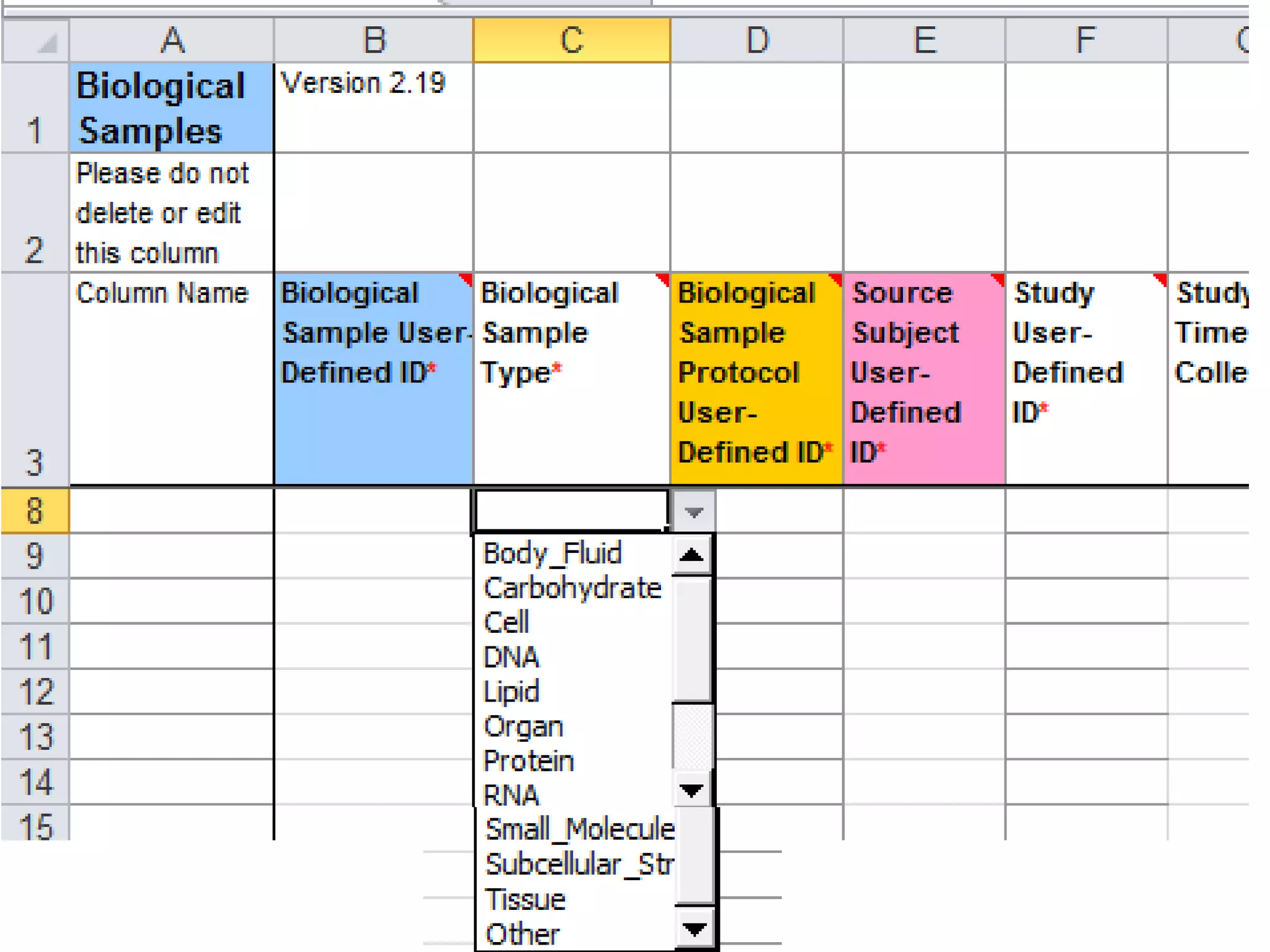
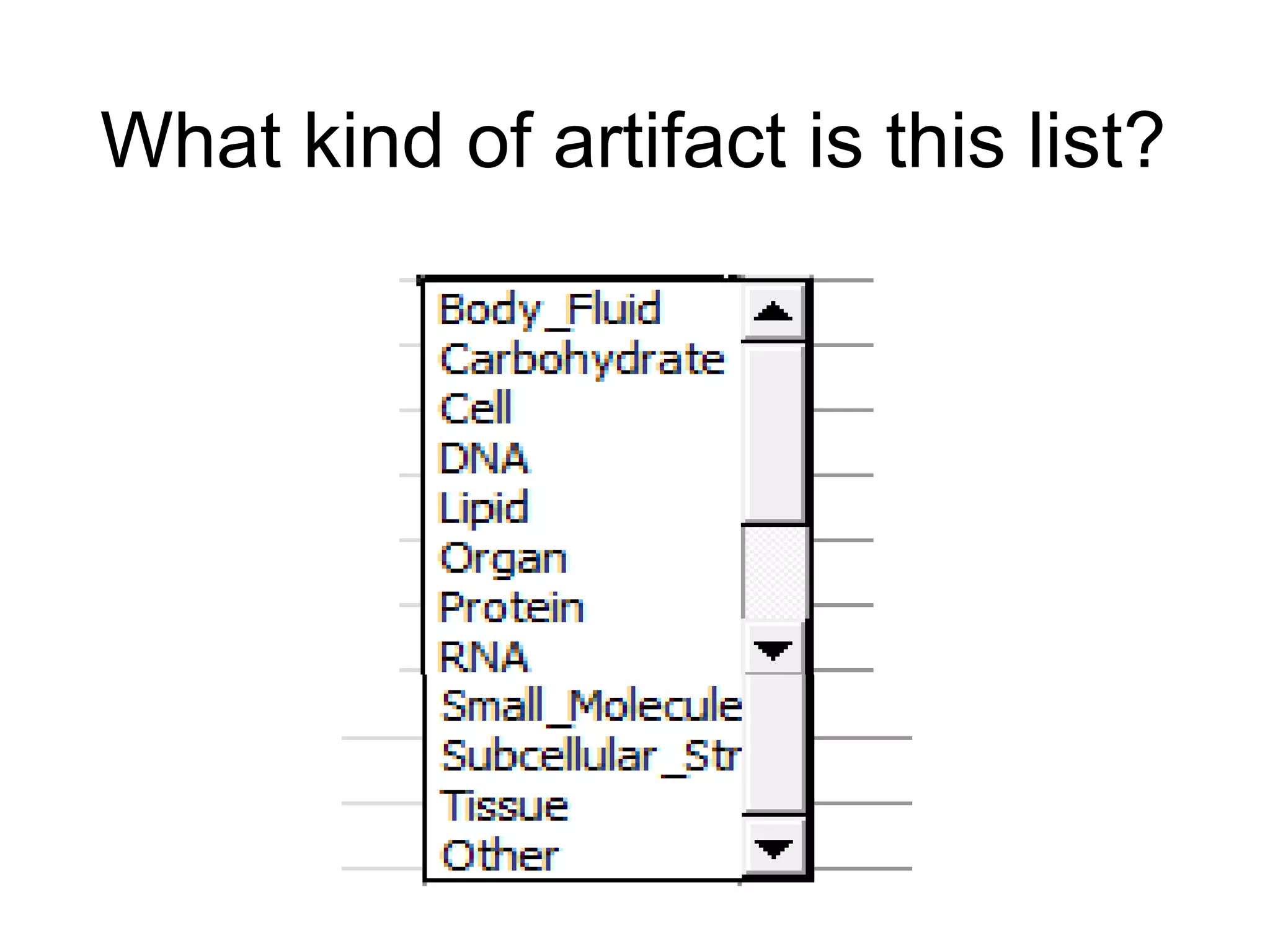
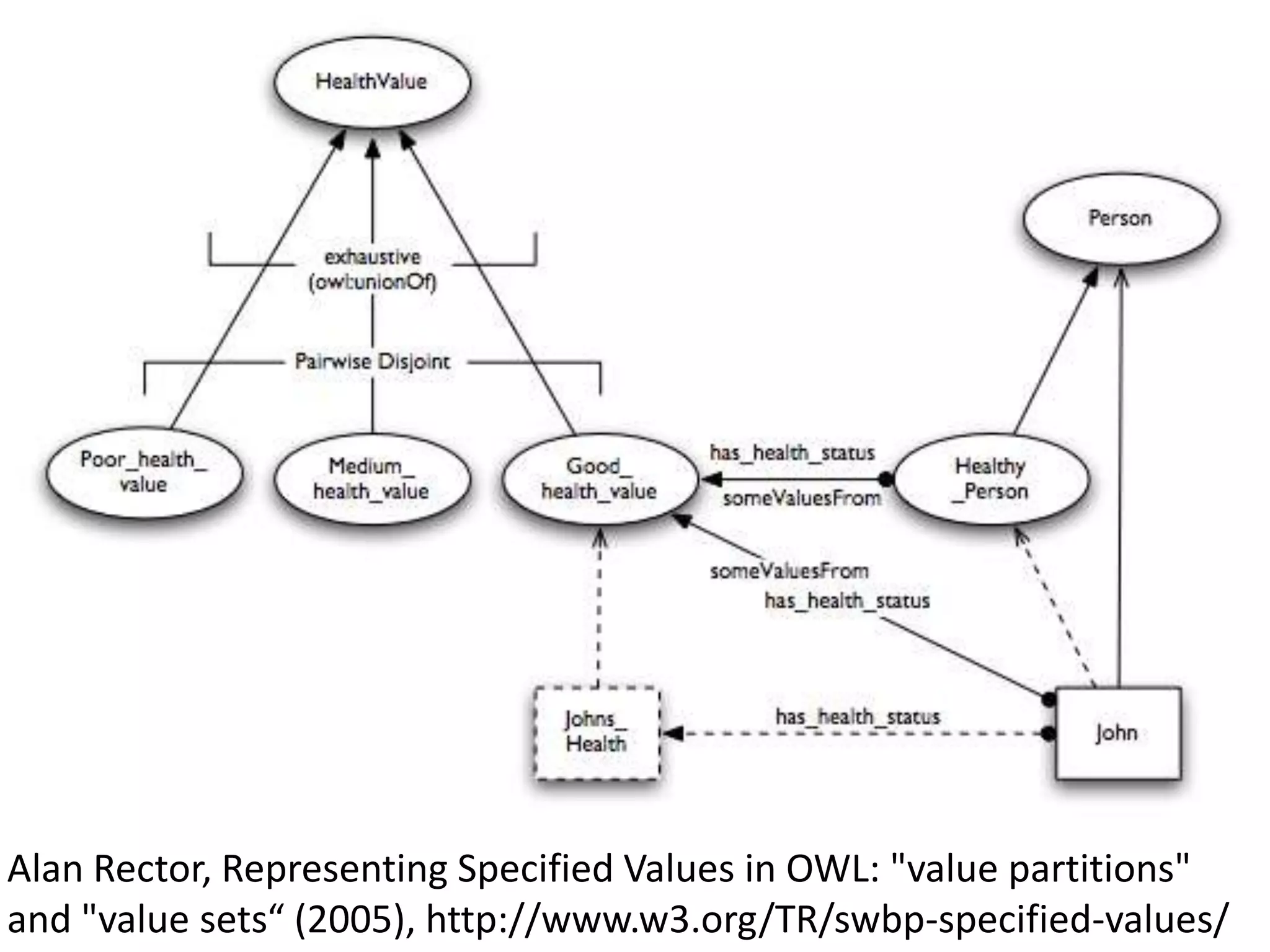
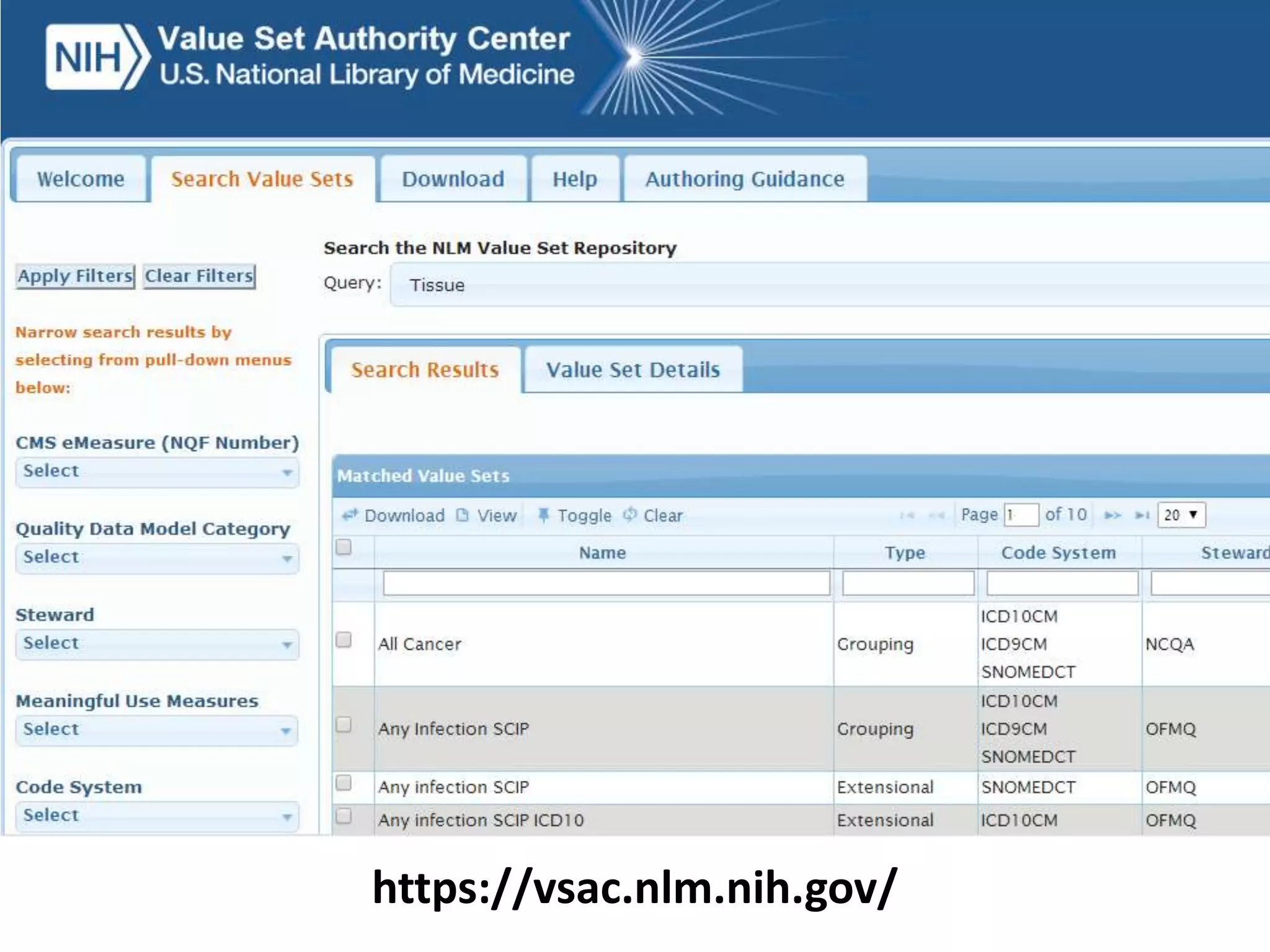
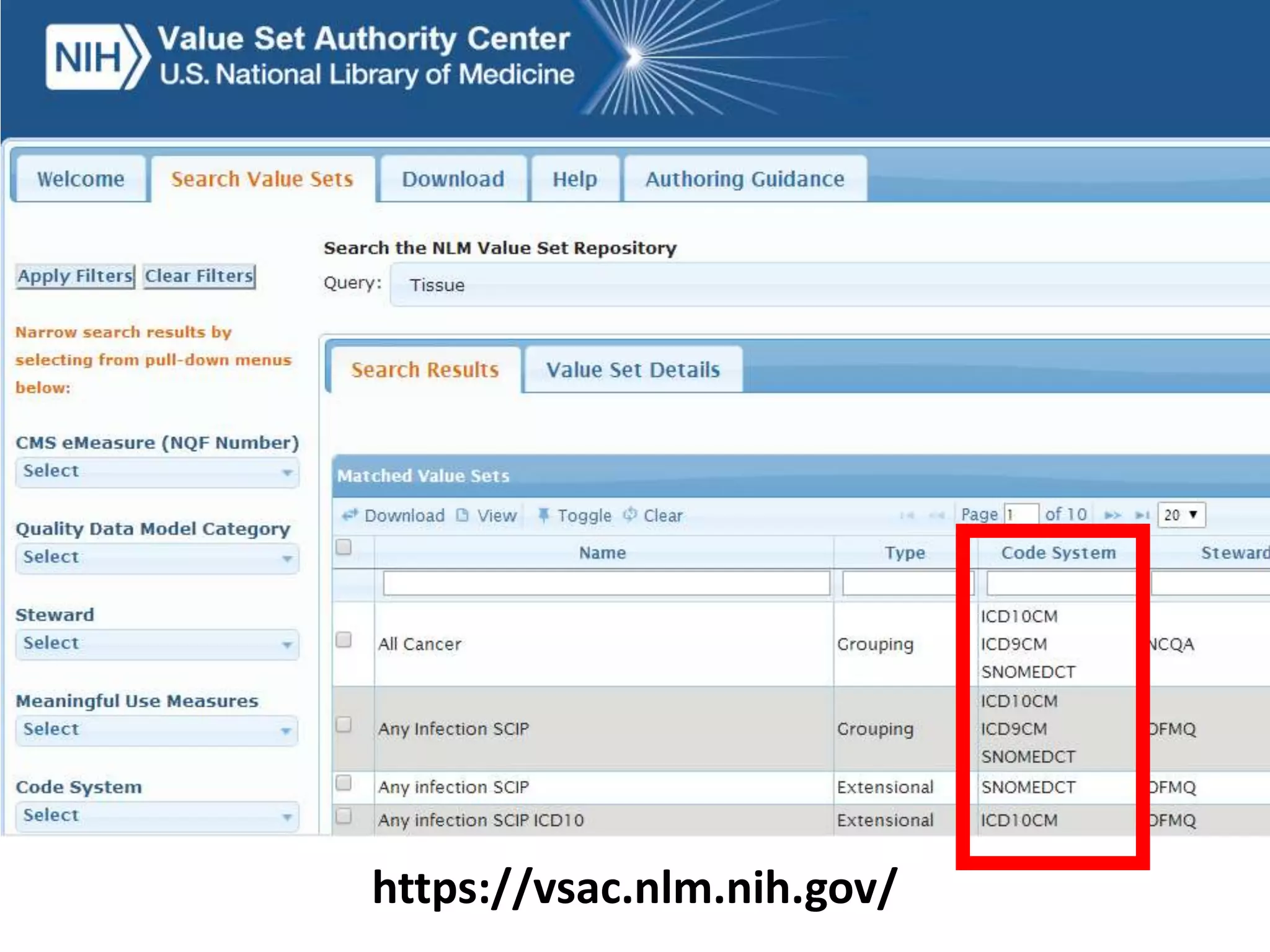
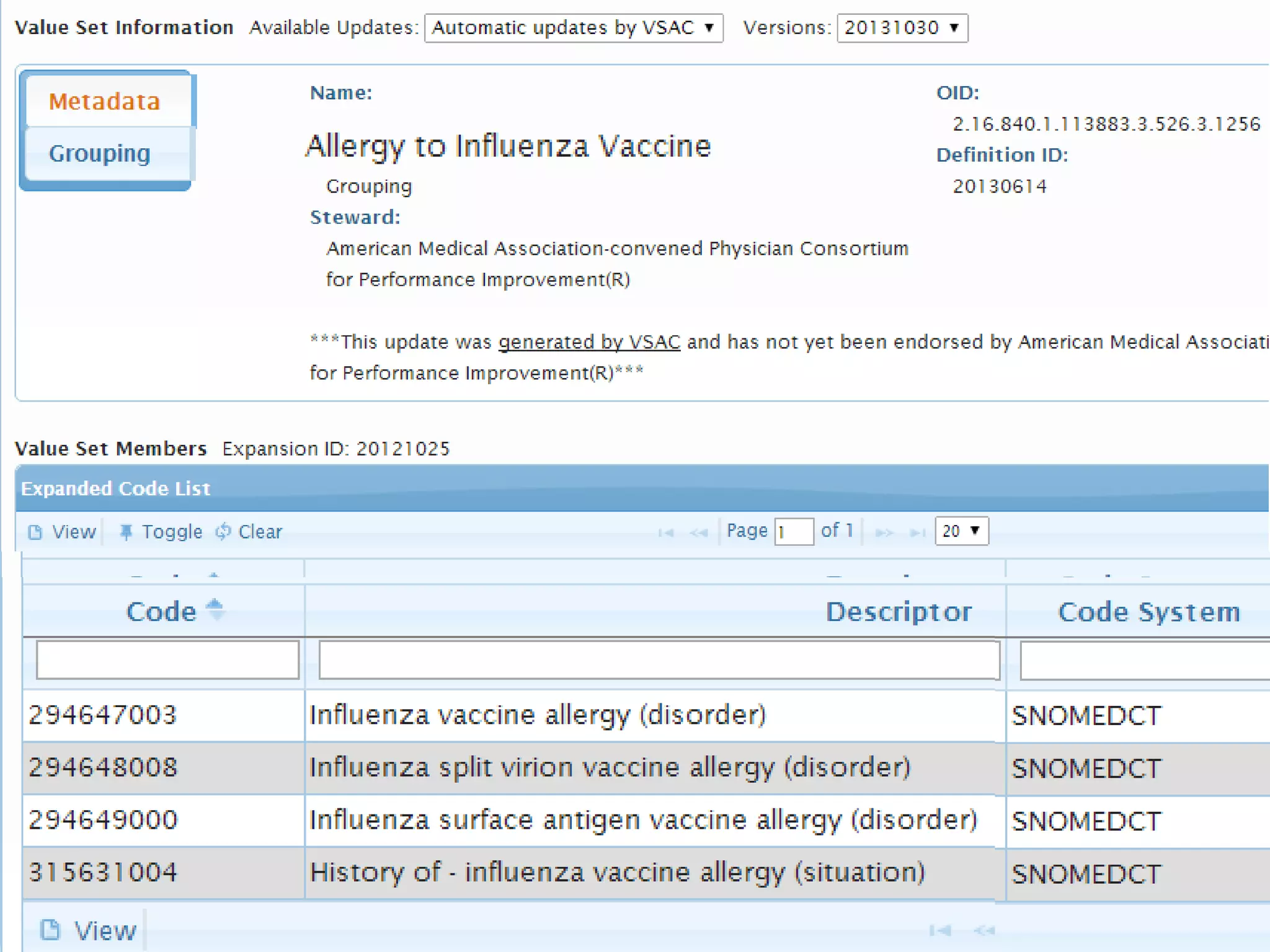
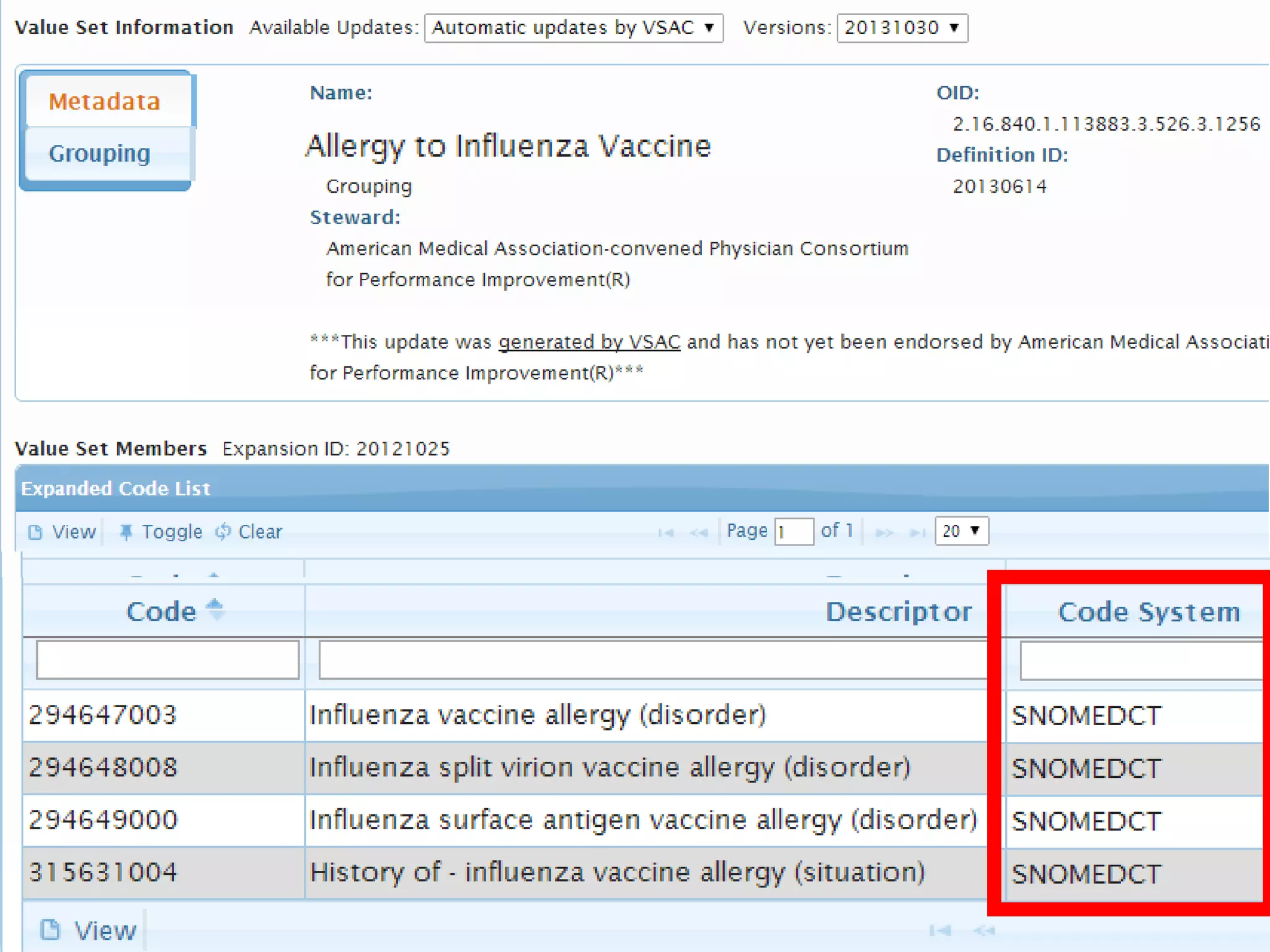
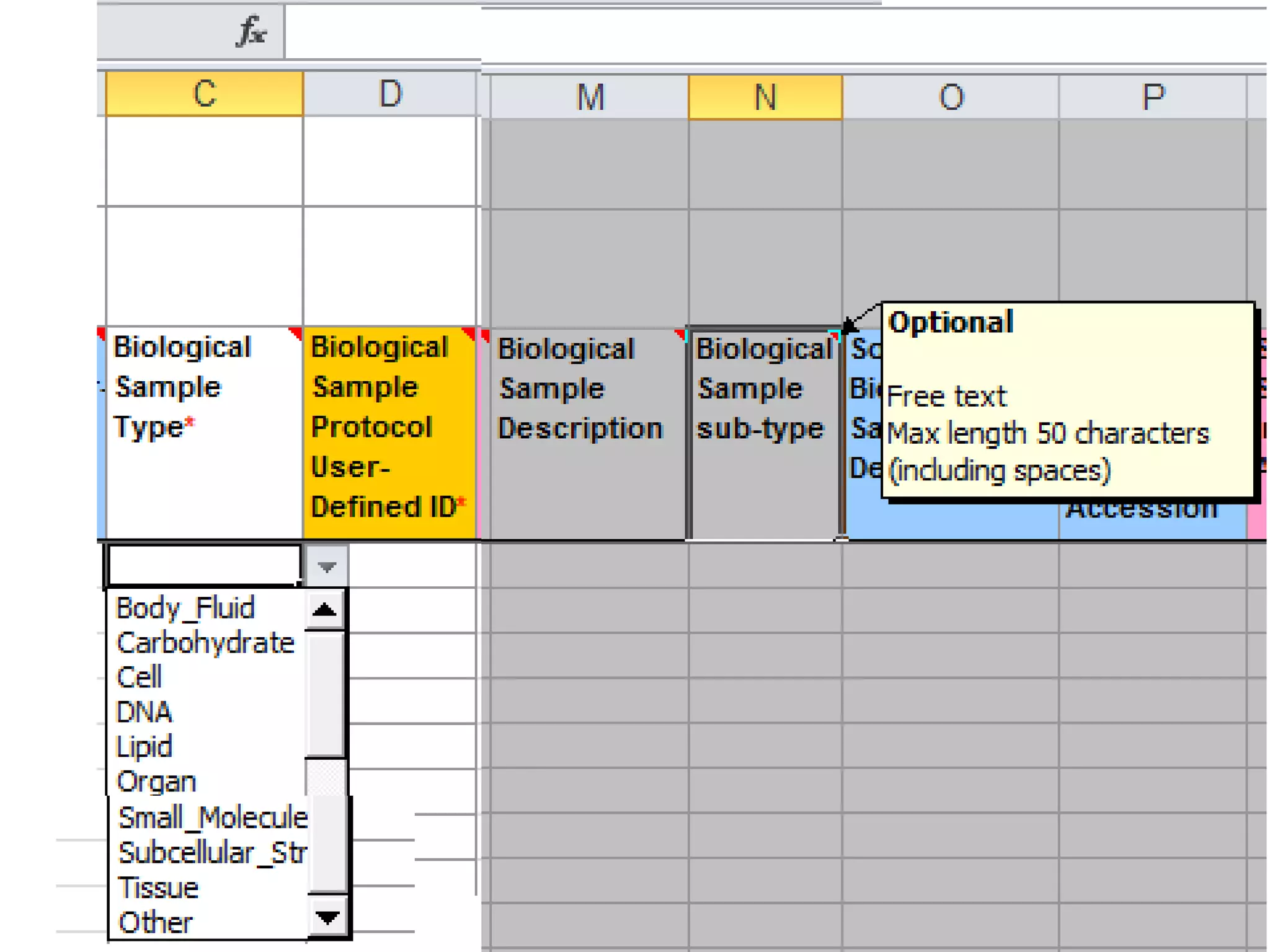
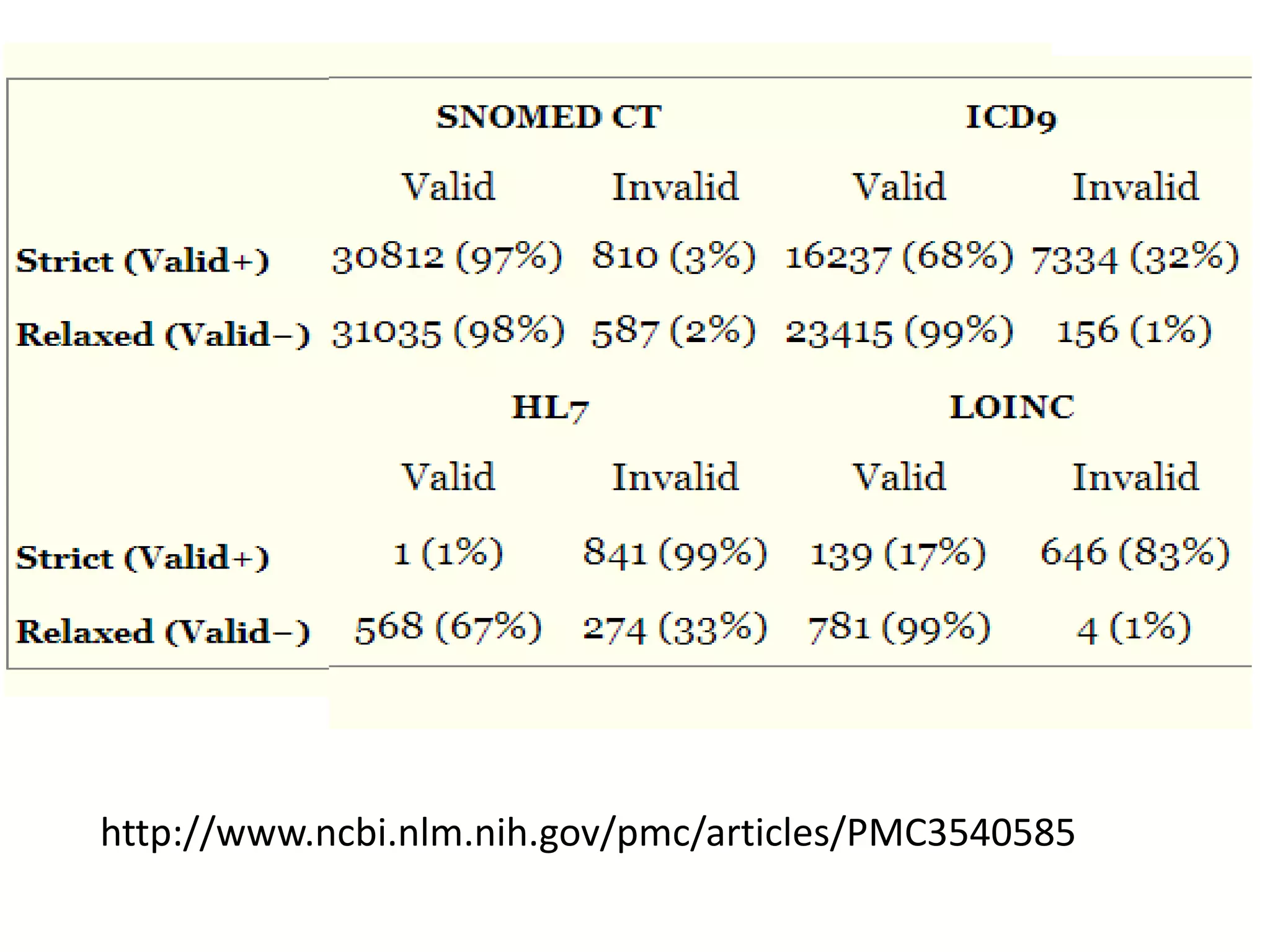
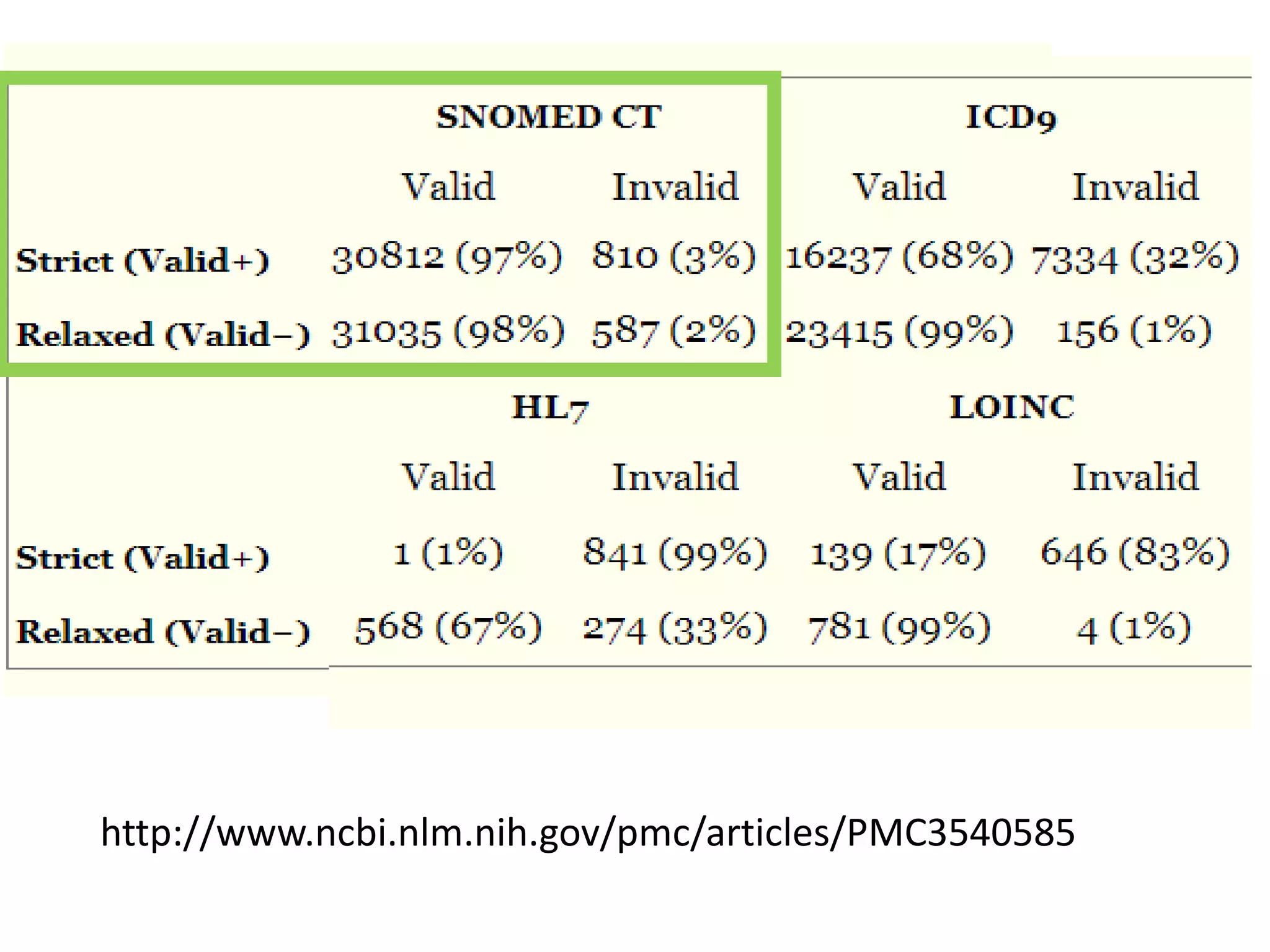
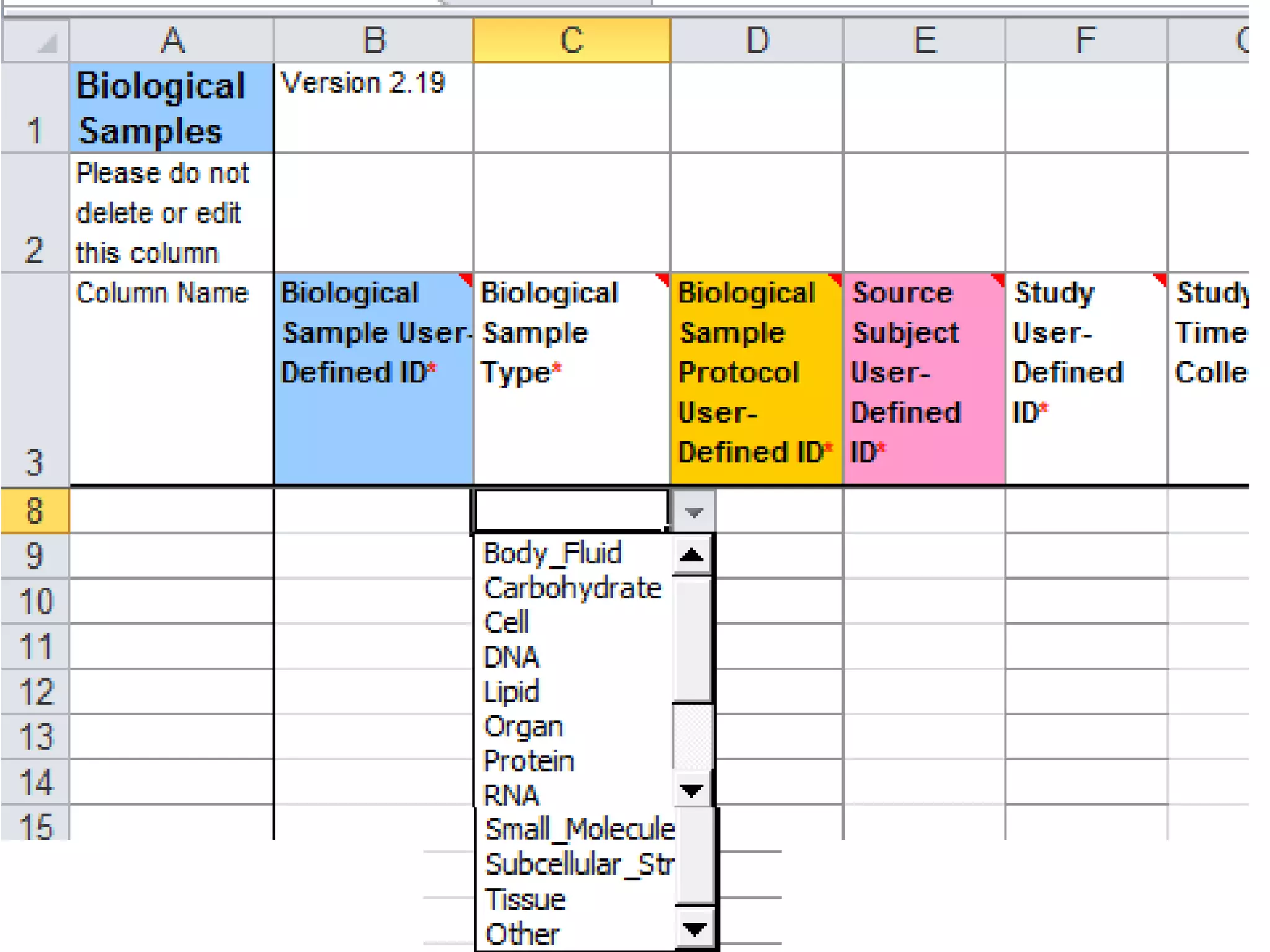
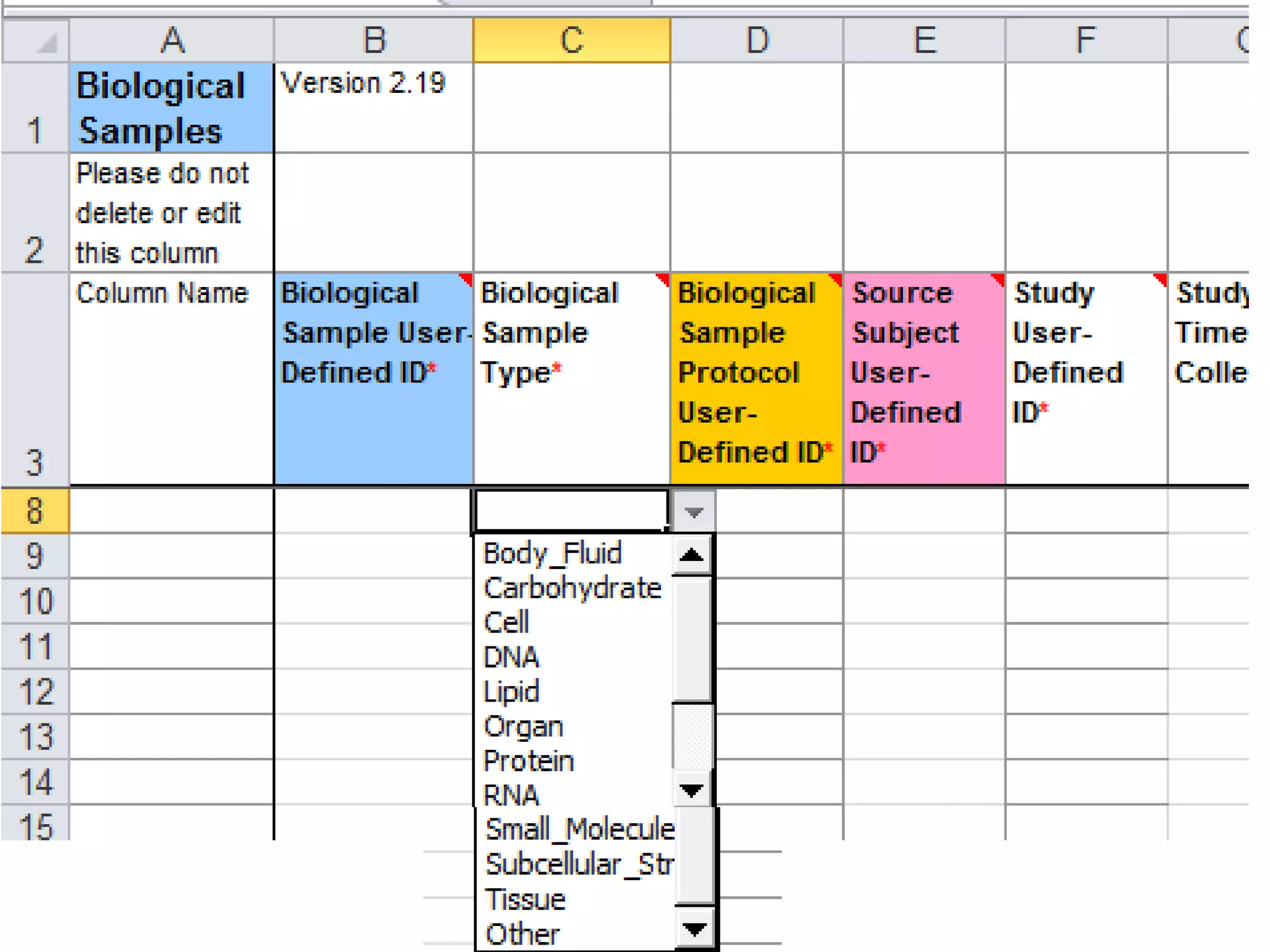
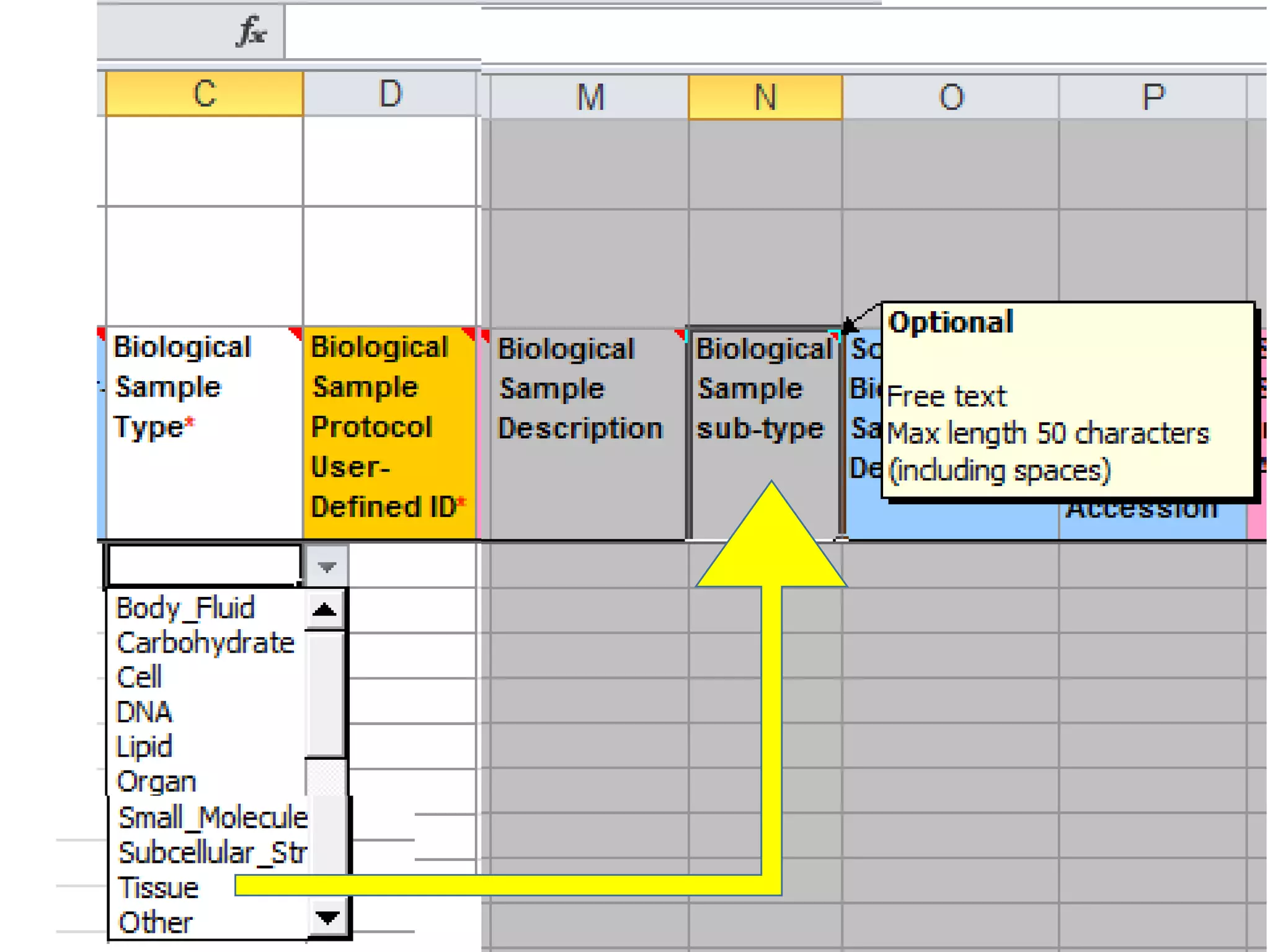
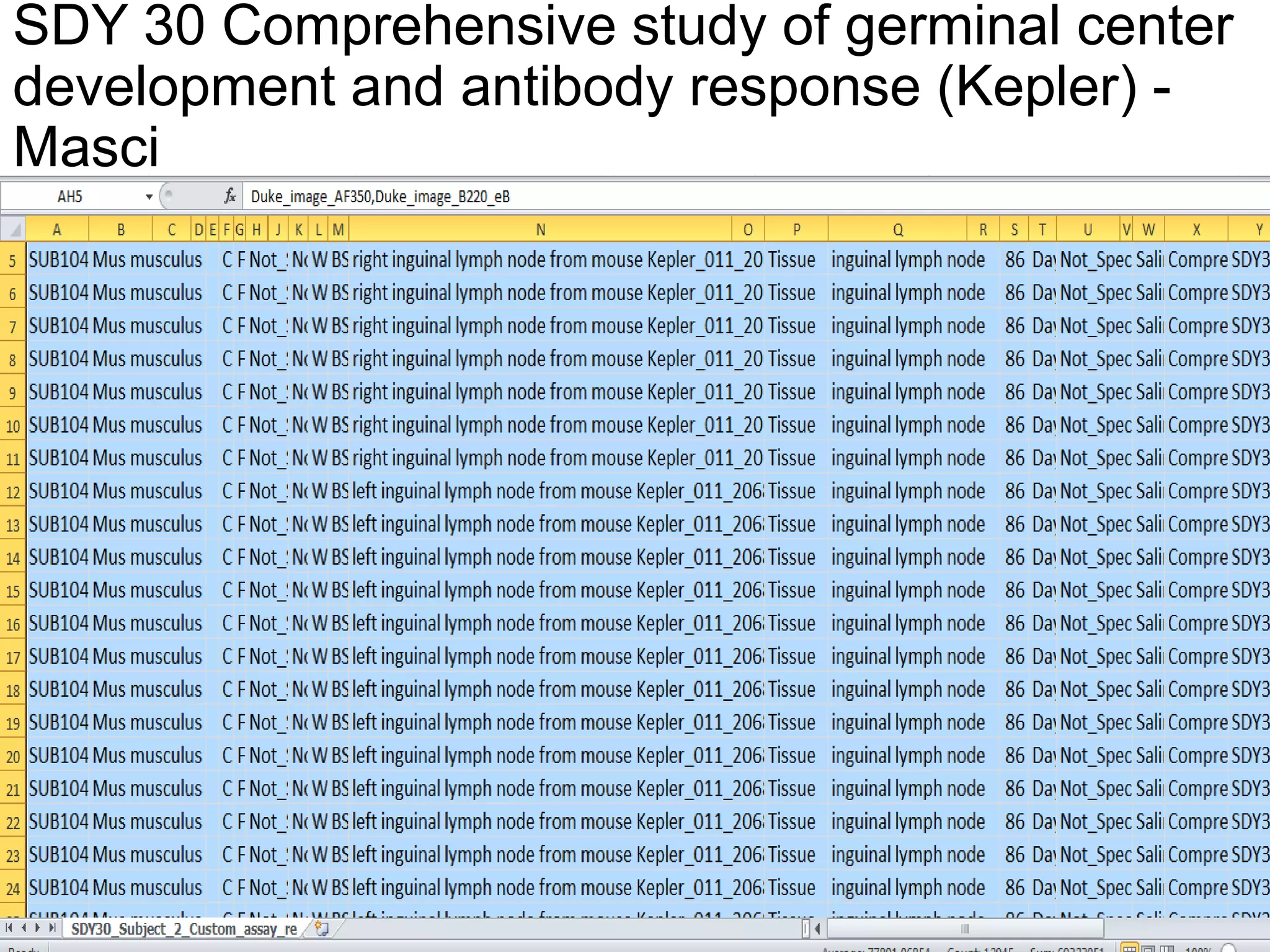
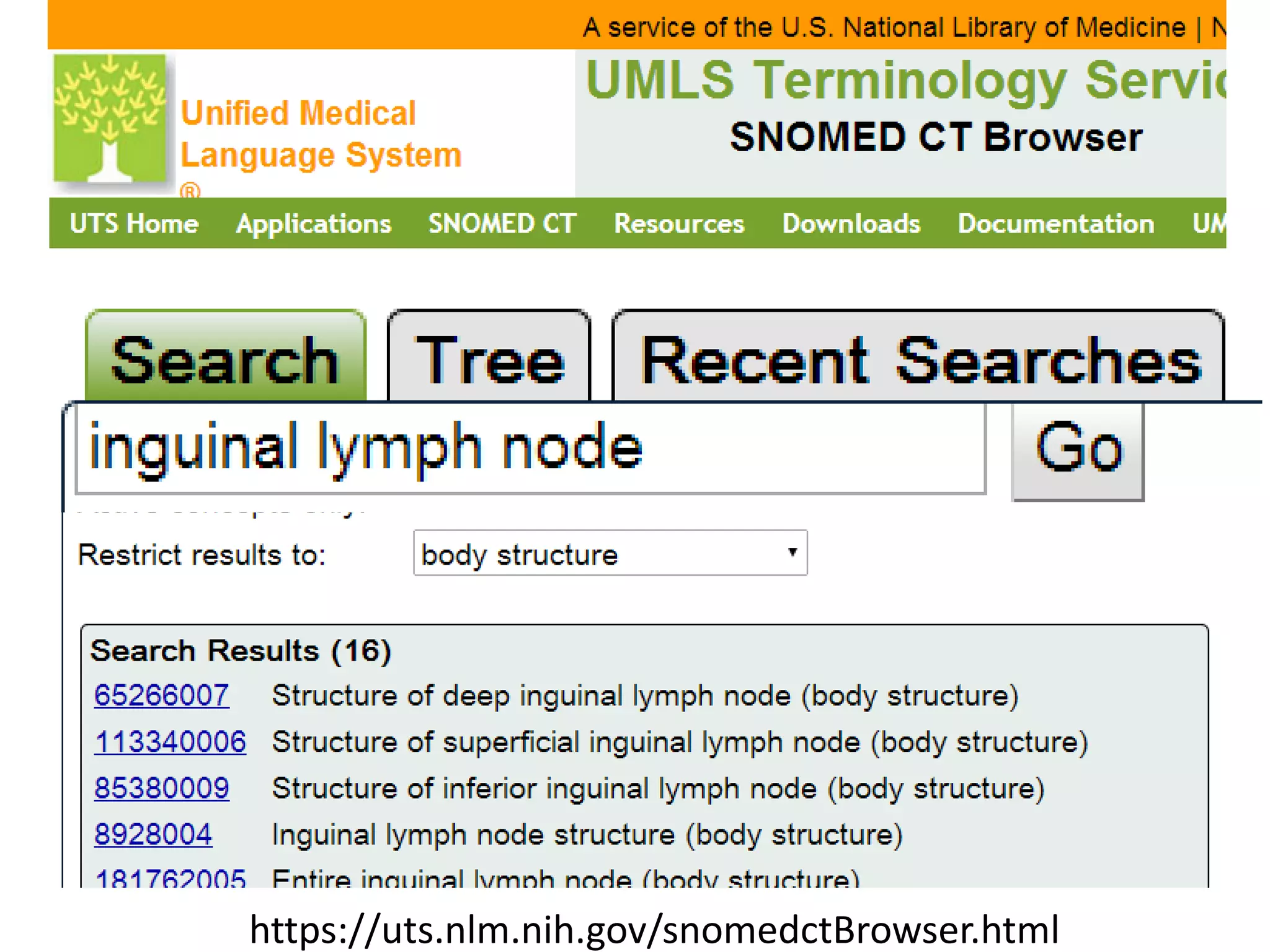
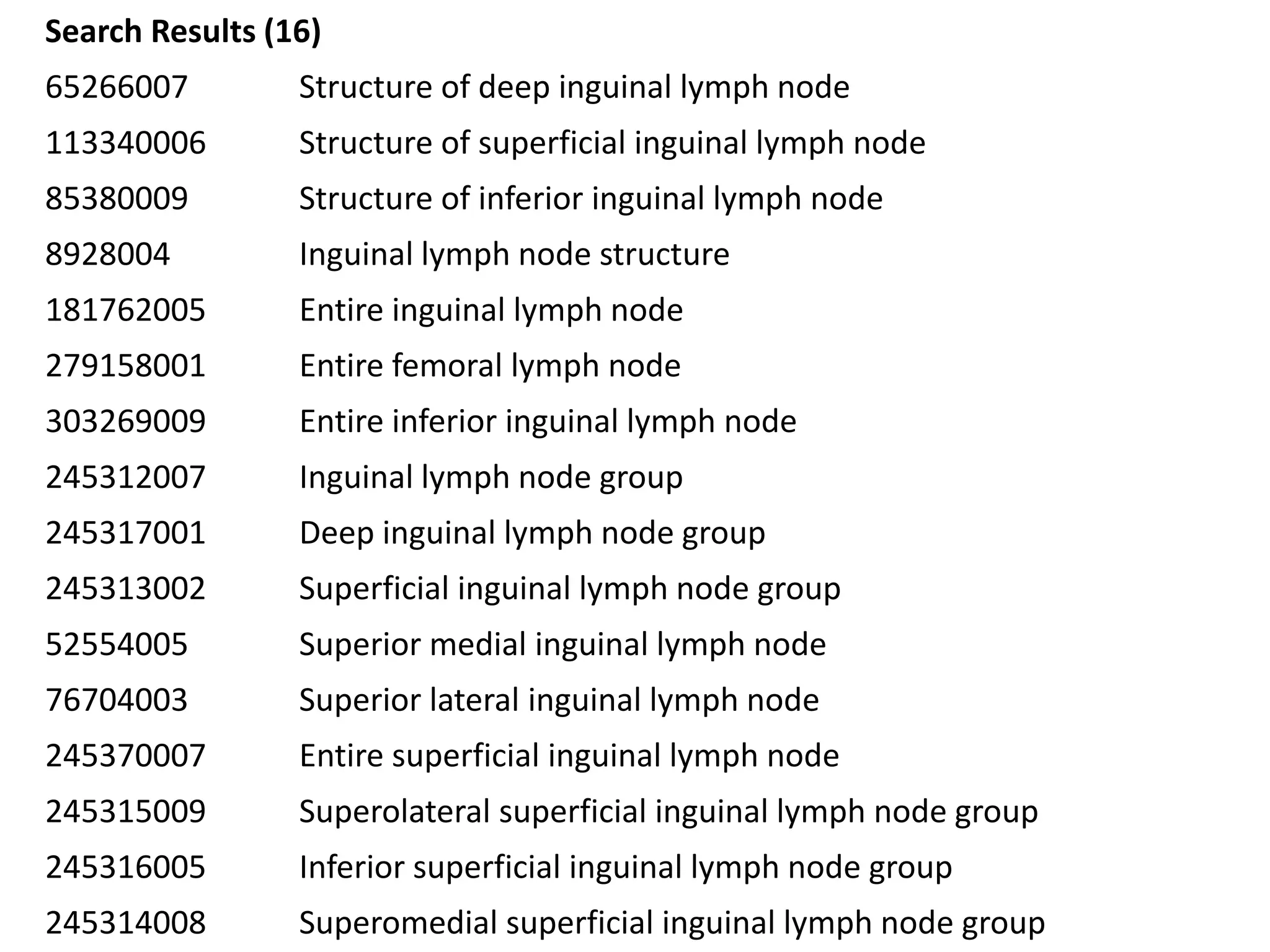
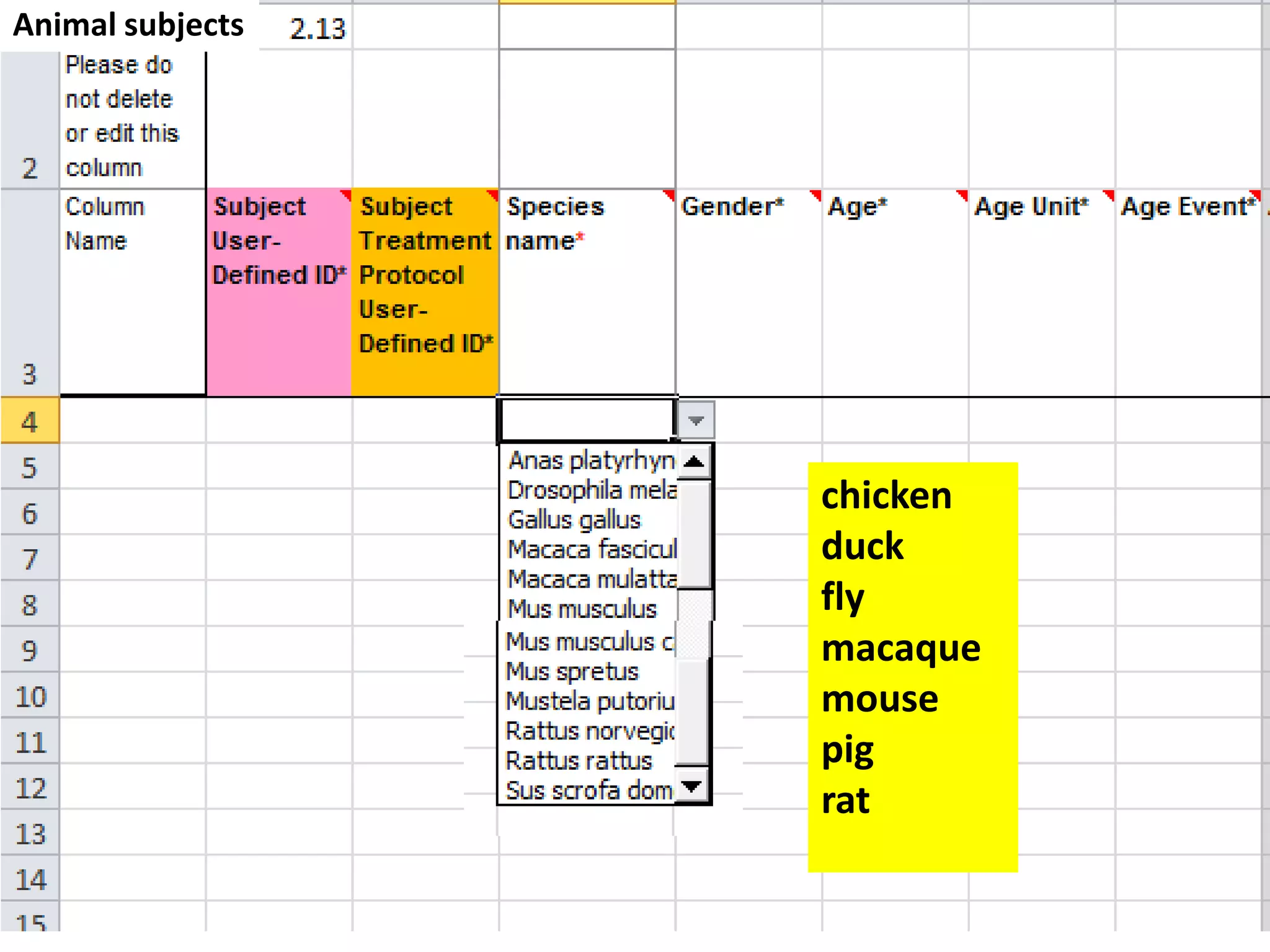
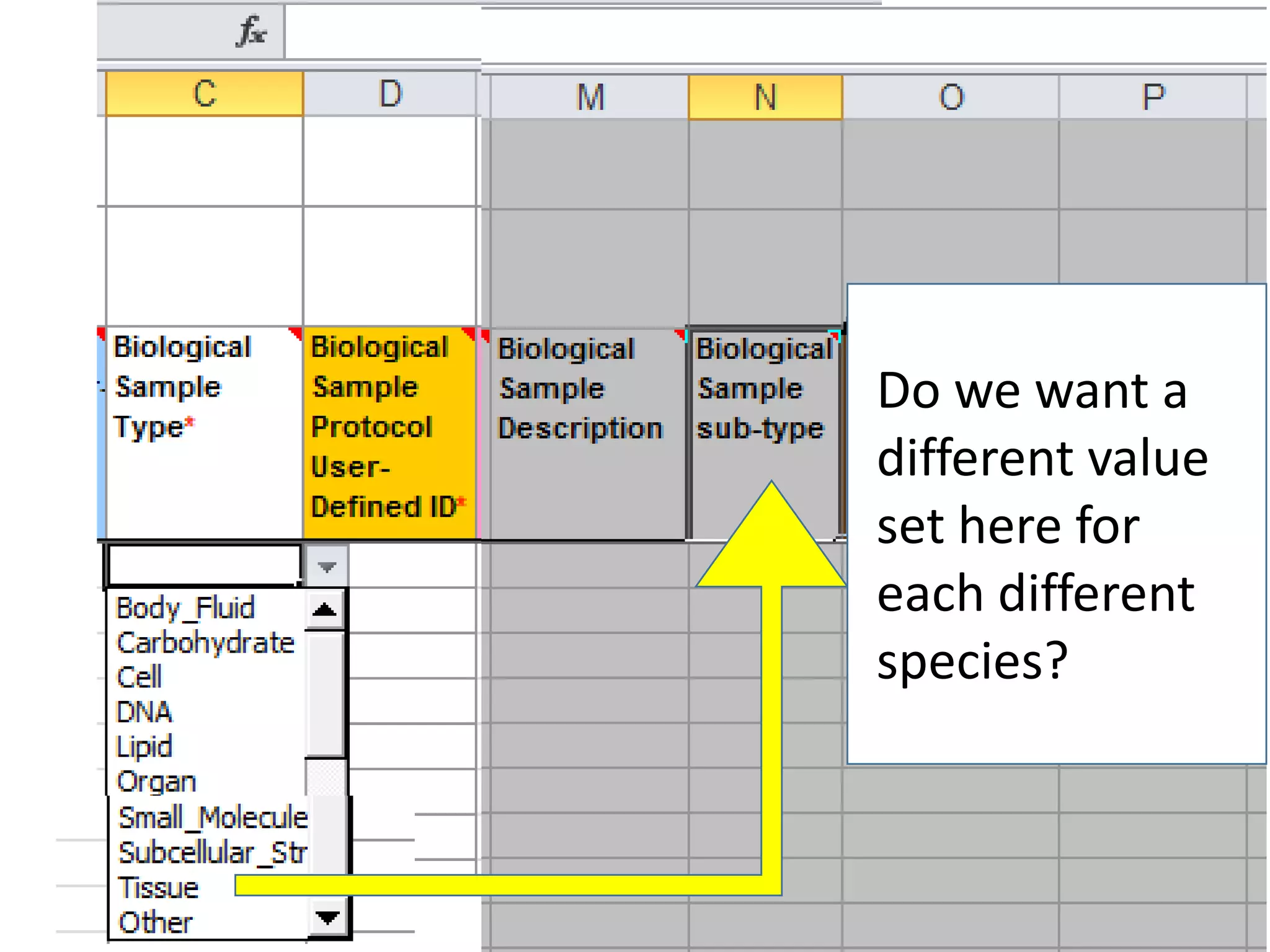
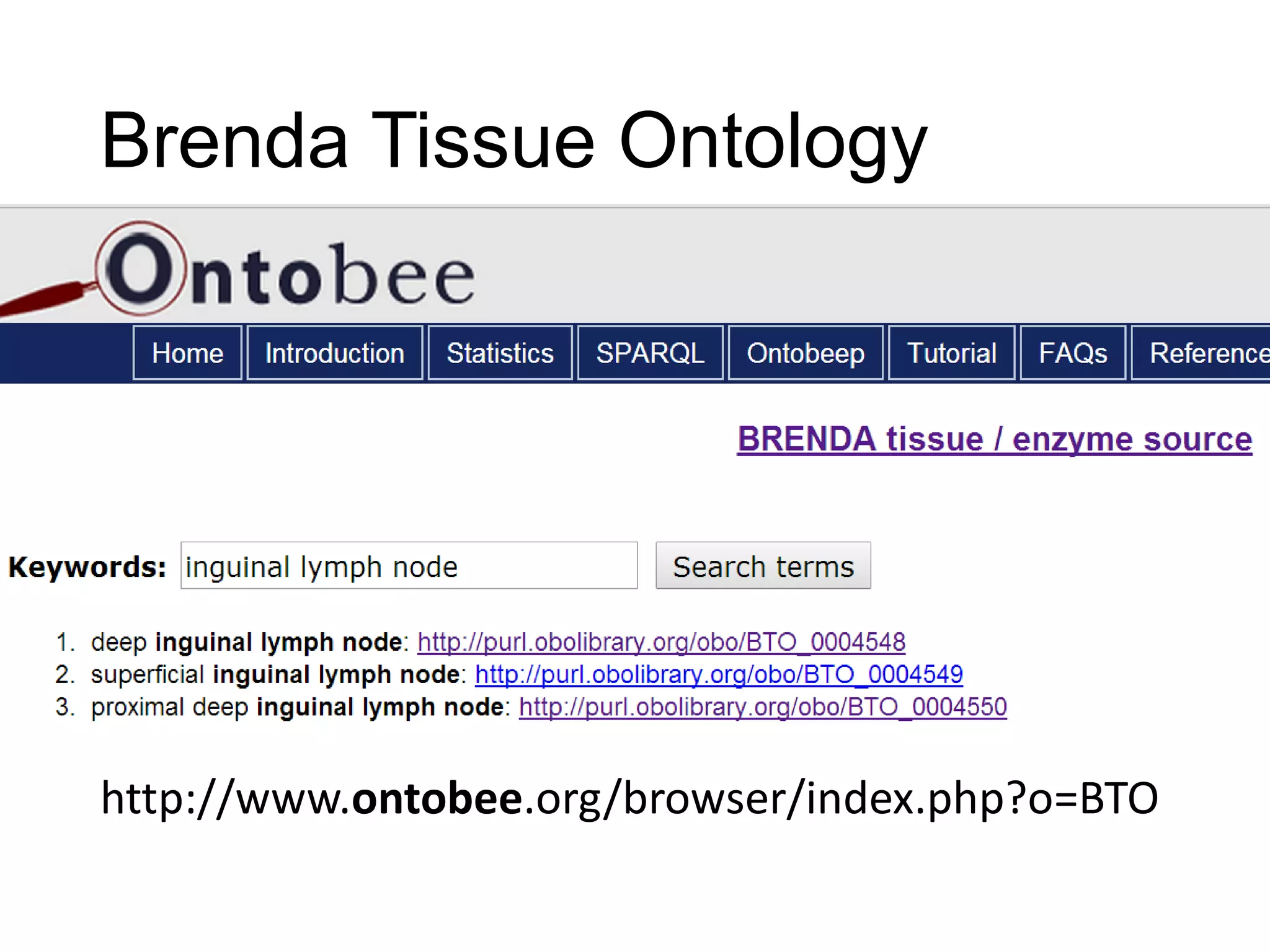
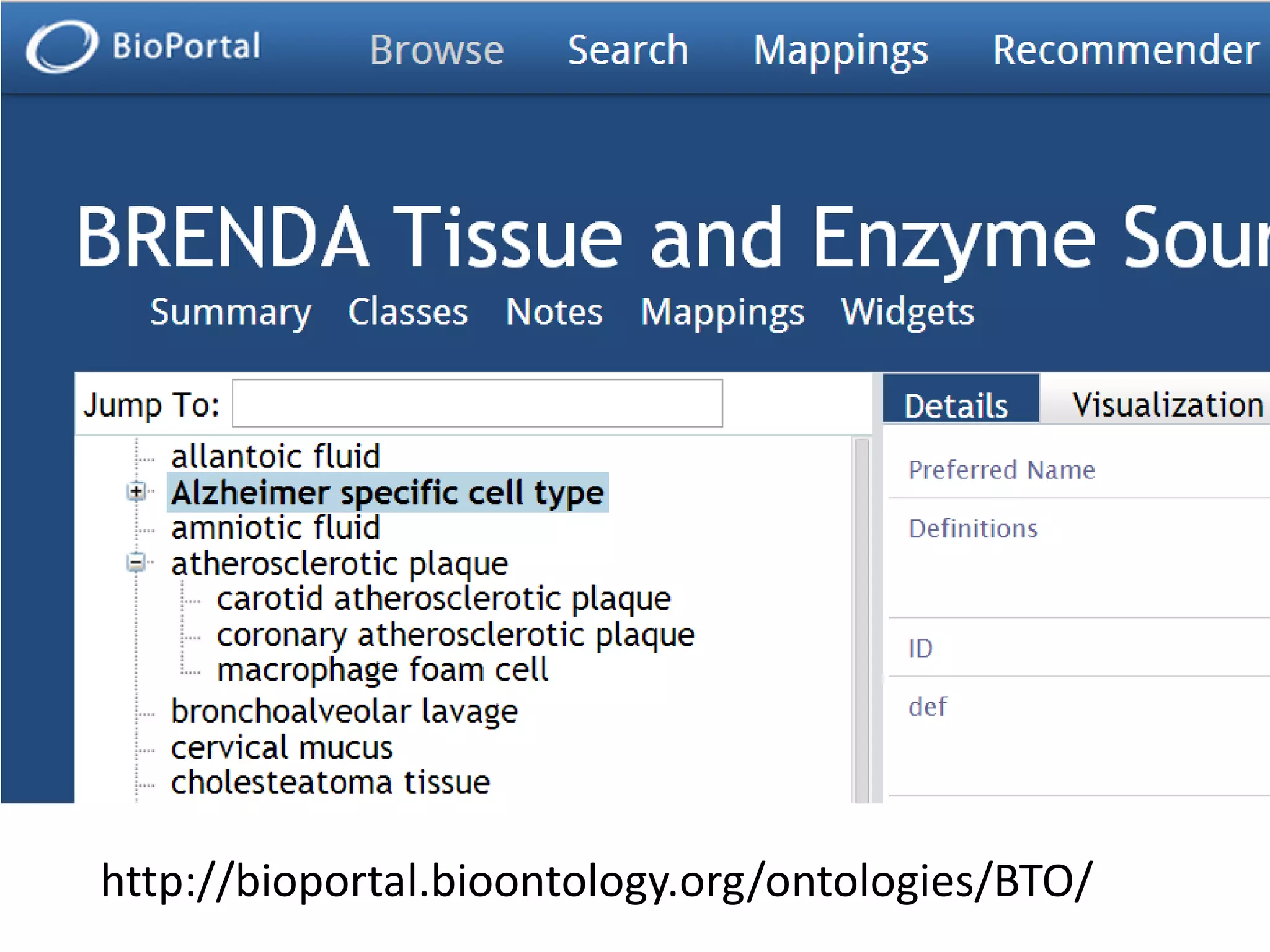
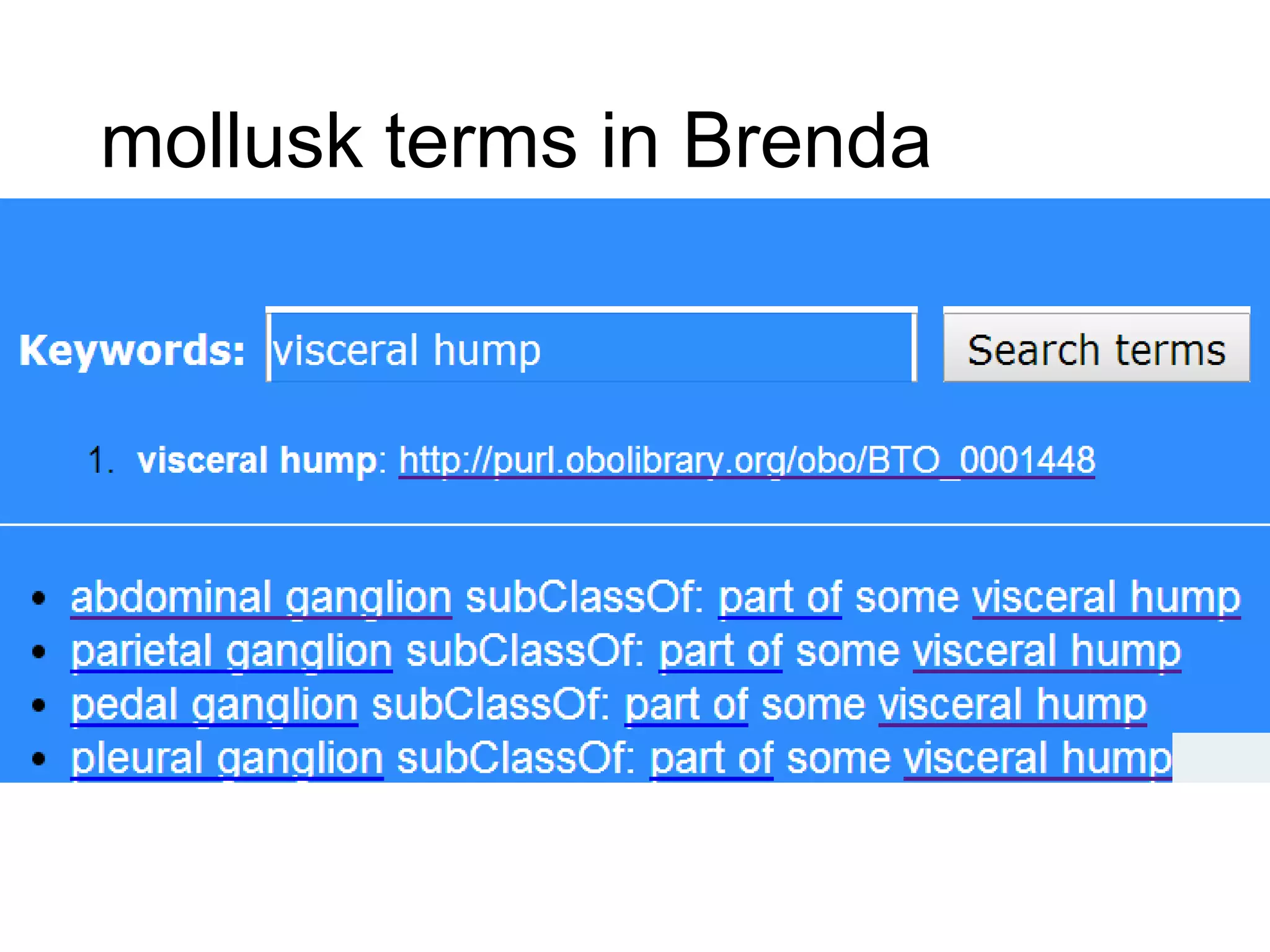
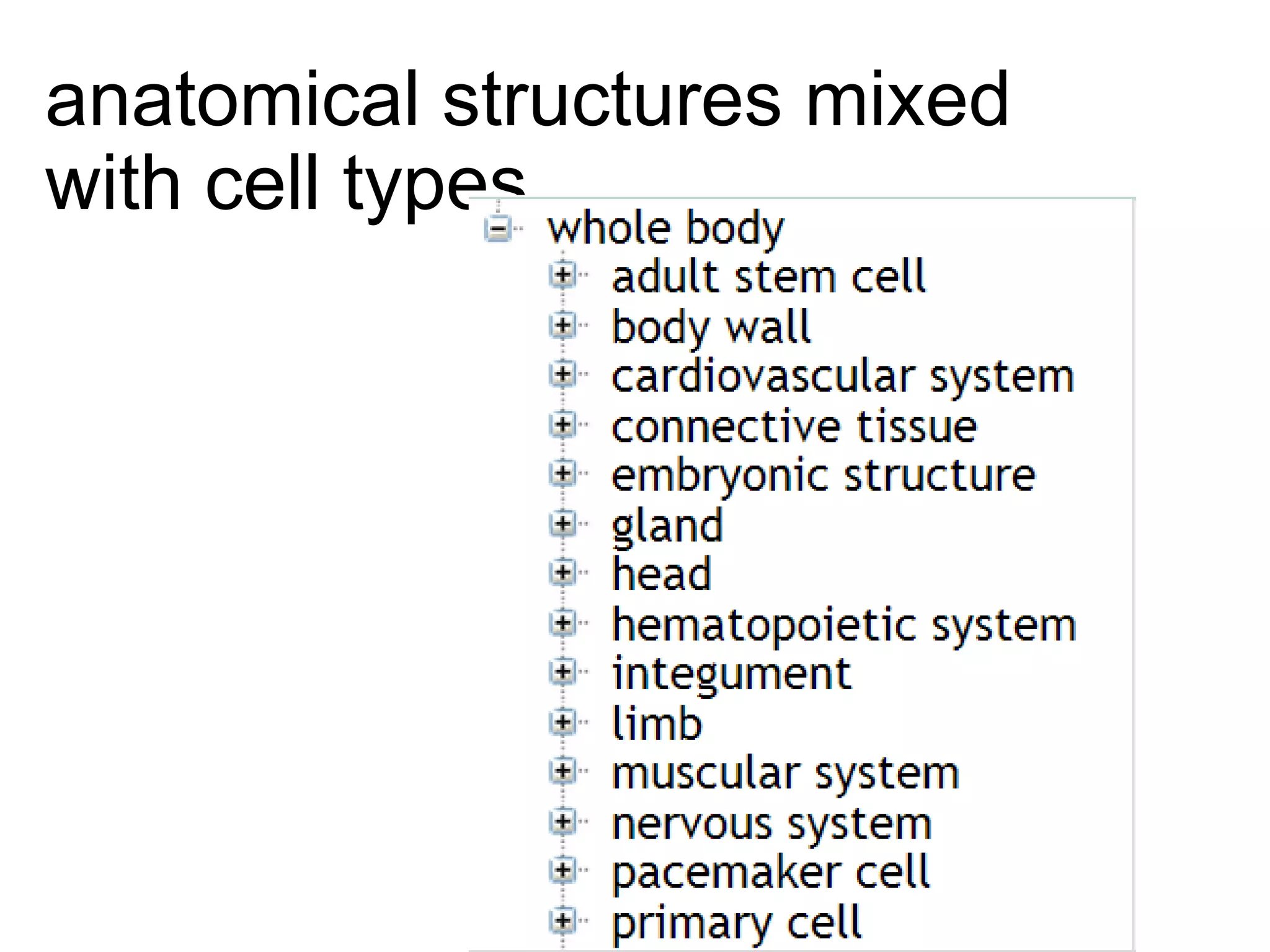
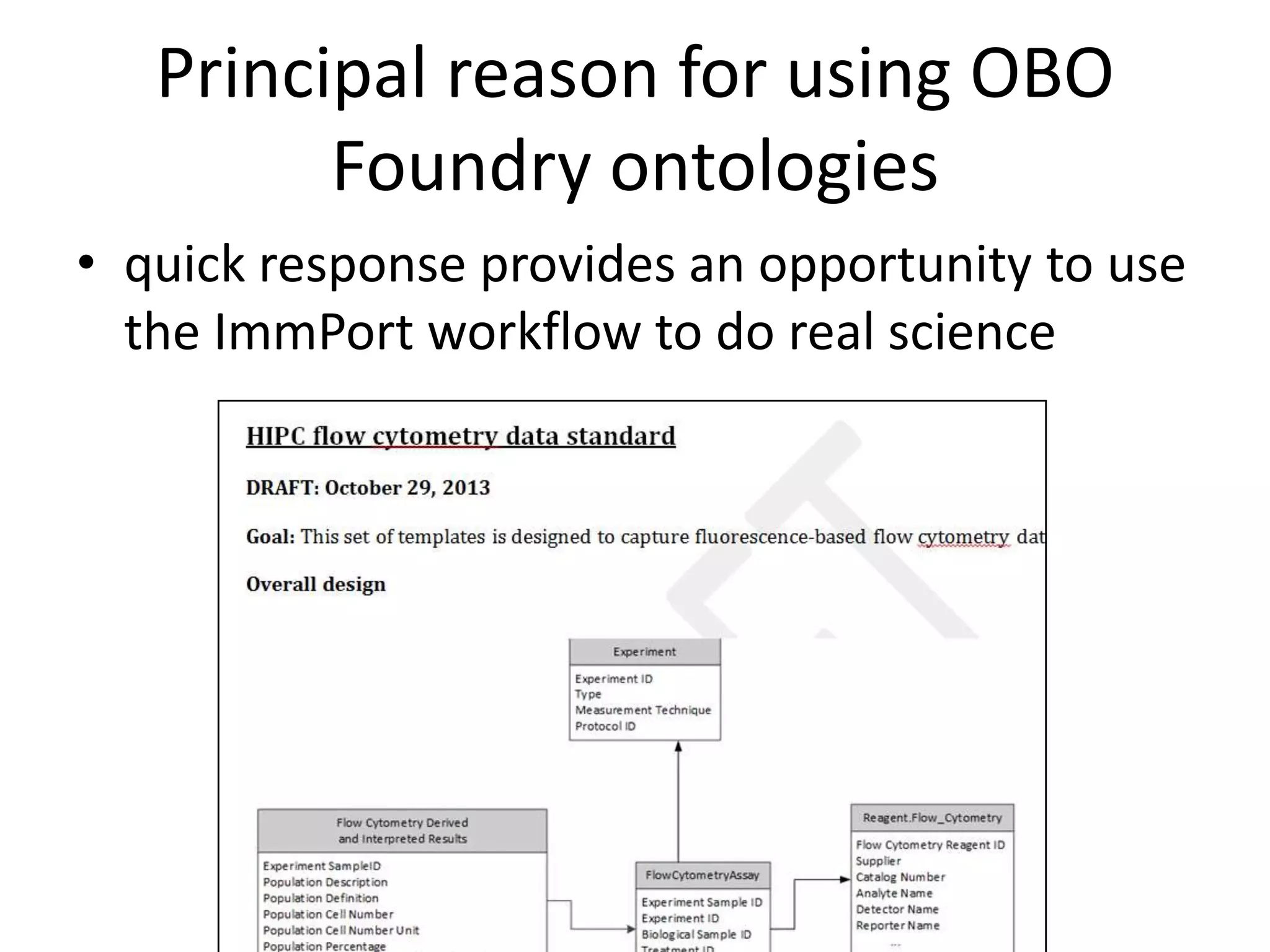
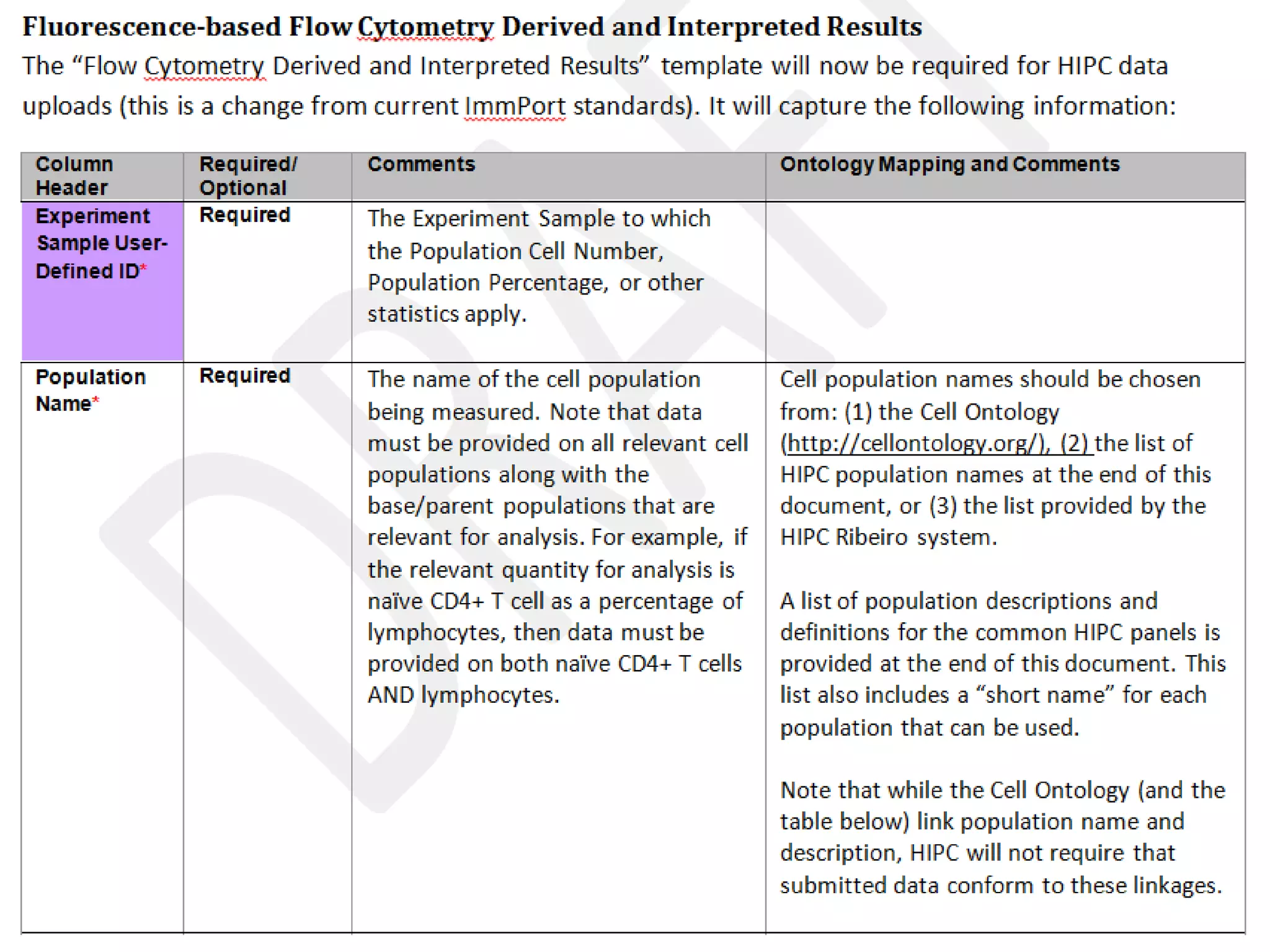
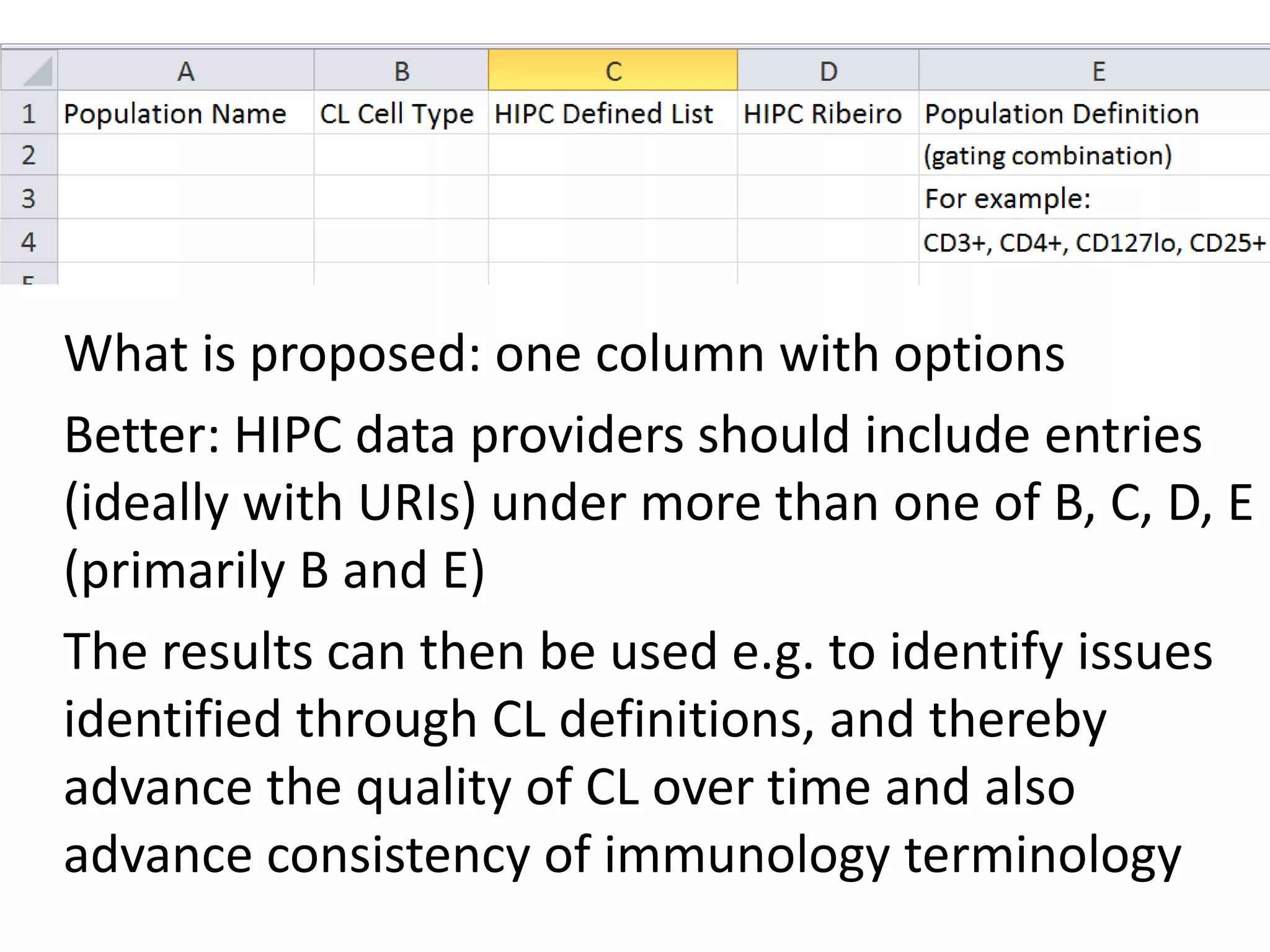
The document discusses enhancing the quality of immport data through the adoption of structured value sets derived from standardized vocabularies and code systems. It highlights the importance of moving away from free text fields to improve data discoverability and comparability, as well as proposing exploring various ontology systems for better categorization and response time. The document also outlines several action items related to evaluating existing coding systems and refining data entry processes for improved quality in research outputs.