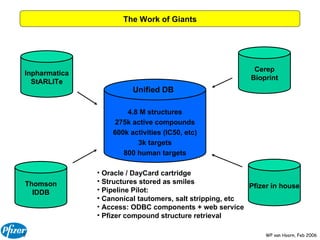
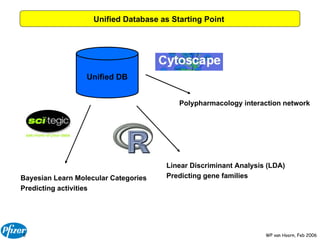
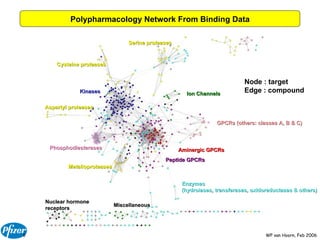
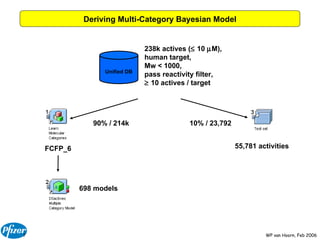
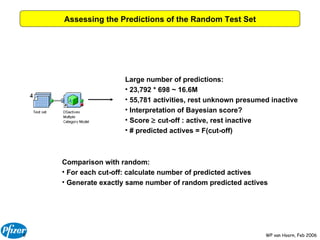
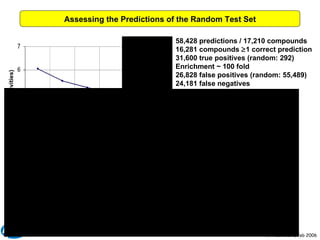
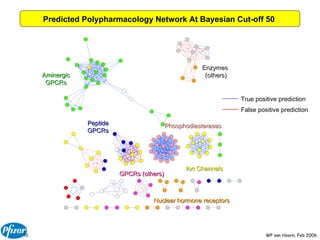
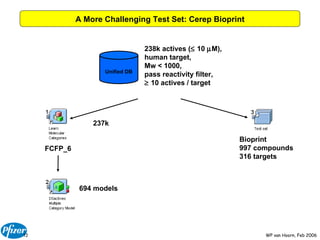
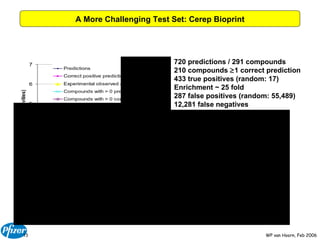
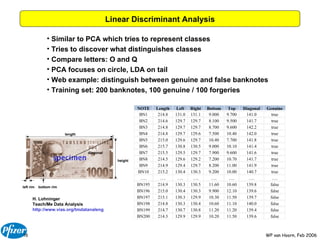
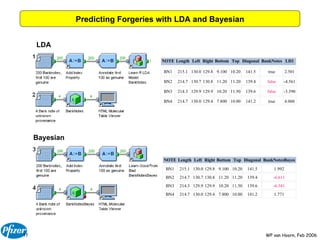
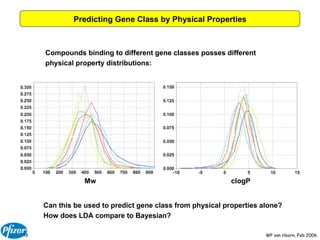
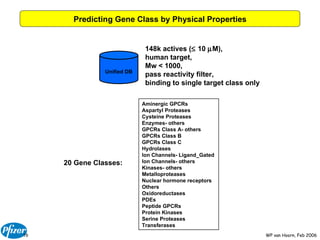
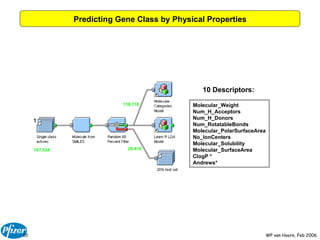
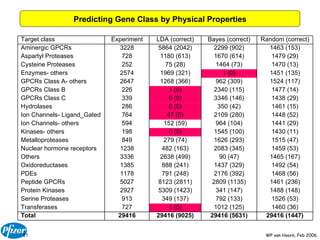
The document discusses the application of Bayesian models and linear discriminant analysis (LDA) in predicting pharmacological activities and gene classes based on compound properties. It highlights the integration of extensive compound datasets, the predictive capabilities of multi-category Bayesian models, and compares their performance with random predictions. The findings suggest that predictive models can significantly enhance screening efficiency and reduce costs in drug design, while also emphasizing the importance of incorporating experimental data for model accuracy.
![Predicting Pharmacology Willem van Hoorn Pfizer Global Research & Development Sandwich UK [email_address] Pipeline Pilot UGM, San Diego, Mar 2006](https://image.slidesharecdn.com/predictingpharmacologyppugm06-100107050354-phpapp01/85/Predicting-Pharmacology-1-320.jpg)