The document discusses several key aspects of evolution including:
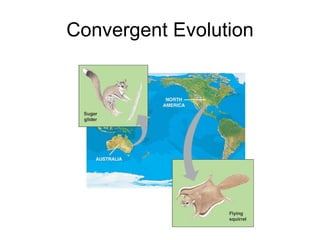
1) Organisms evolve through random mutations that are influenced by environmental factors, with beneficial mutations increasing an organism's chances of survival and reproduction.
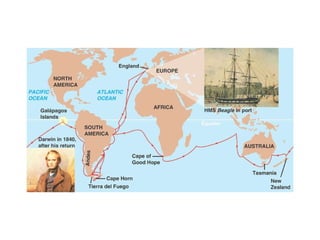
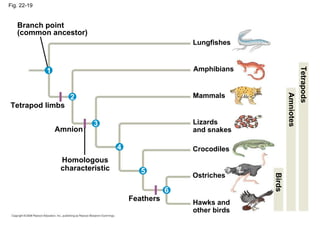
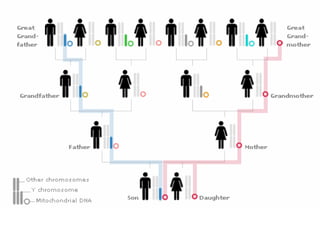
2) Genetic drift, the random changes in allele frequencies within a population from one generation to the next, can cause populations to diverge over time leading to speciation.
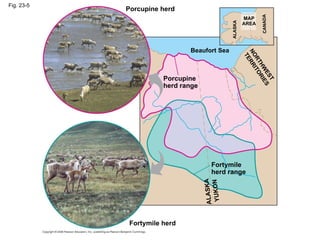
3) Reproductive isolation through mechanisms like habitat, behavioral, and temporal isolation can further drive speciation when populations become separated and diverge genetically.