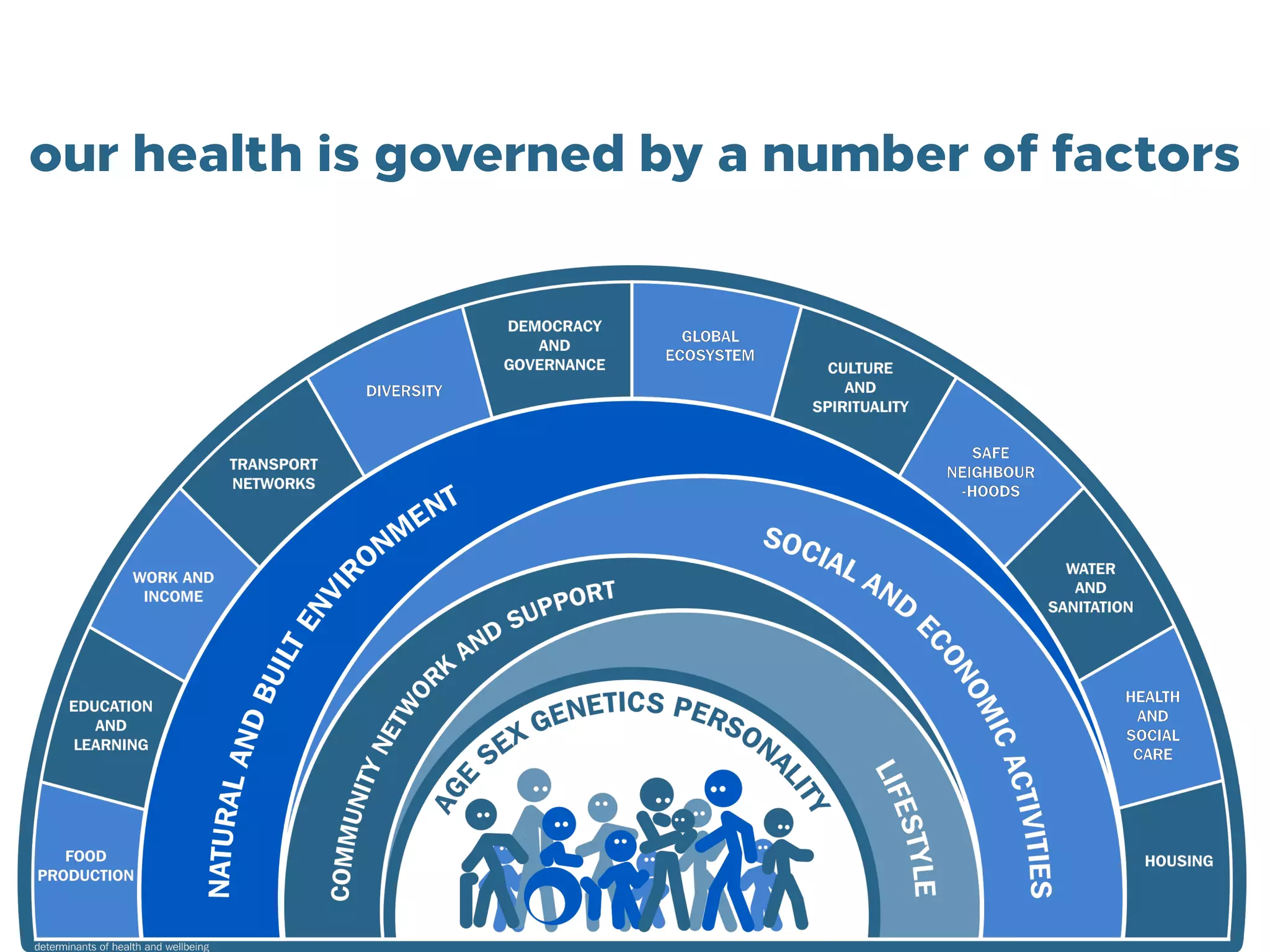
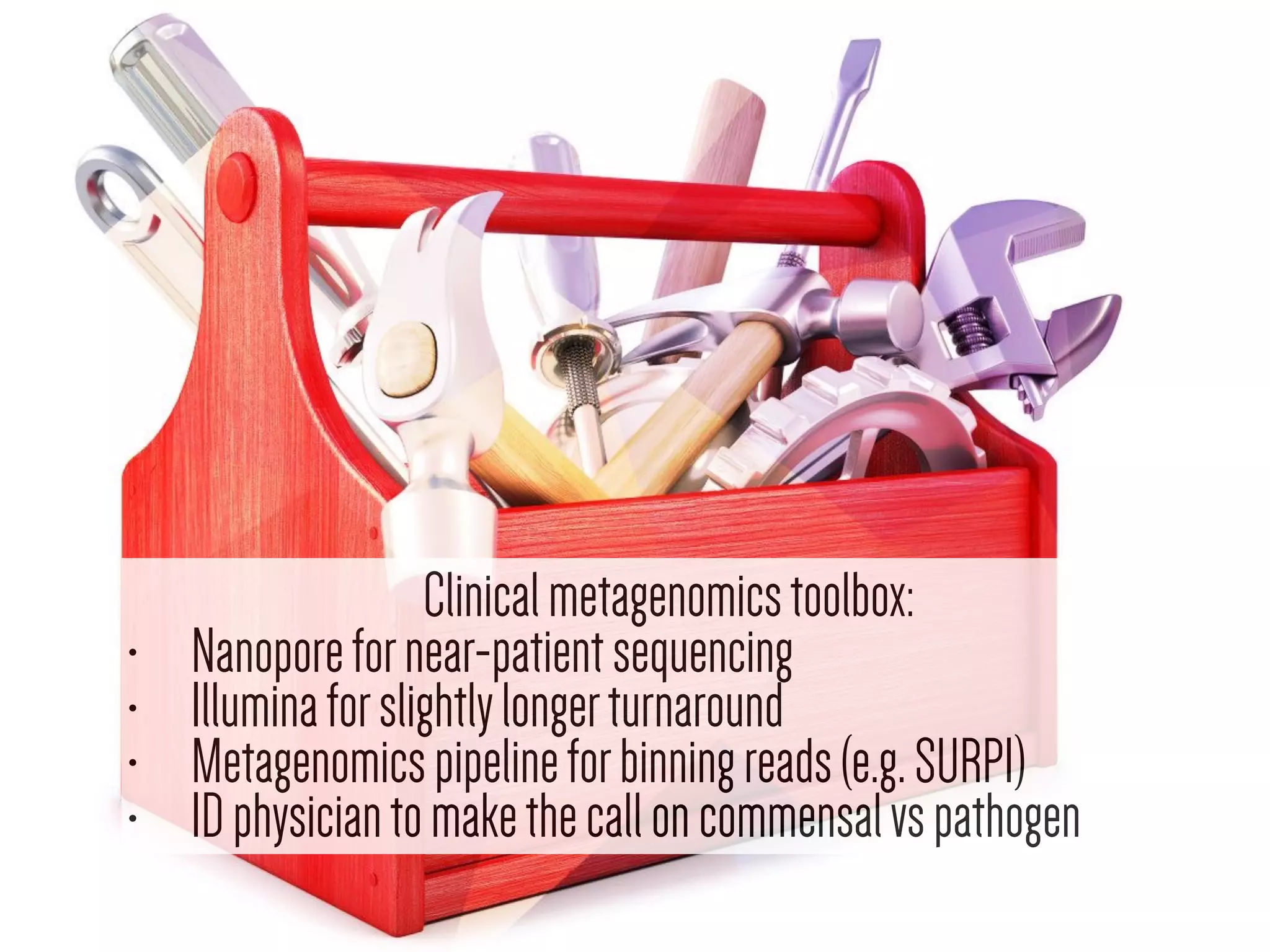
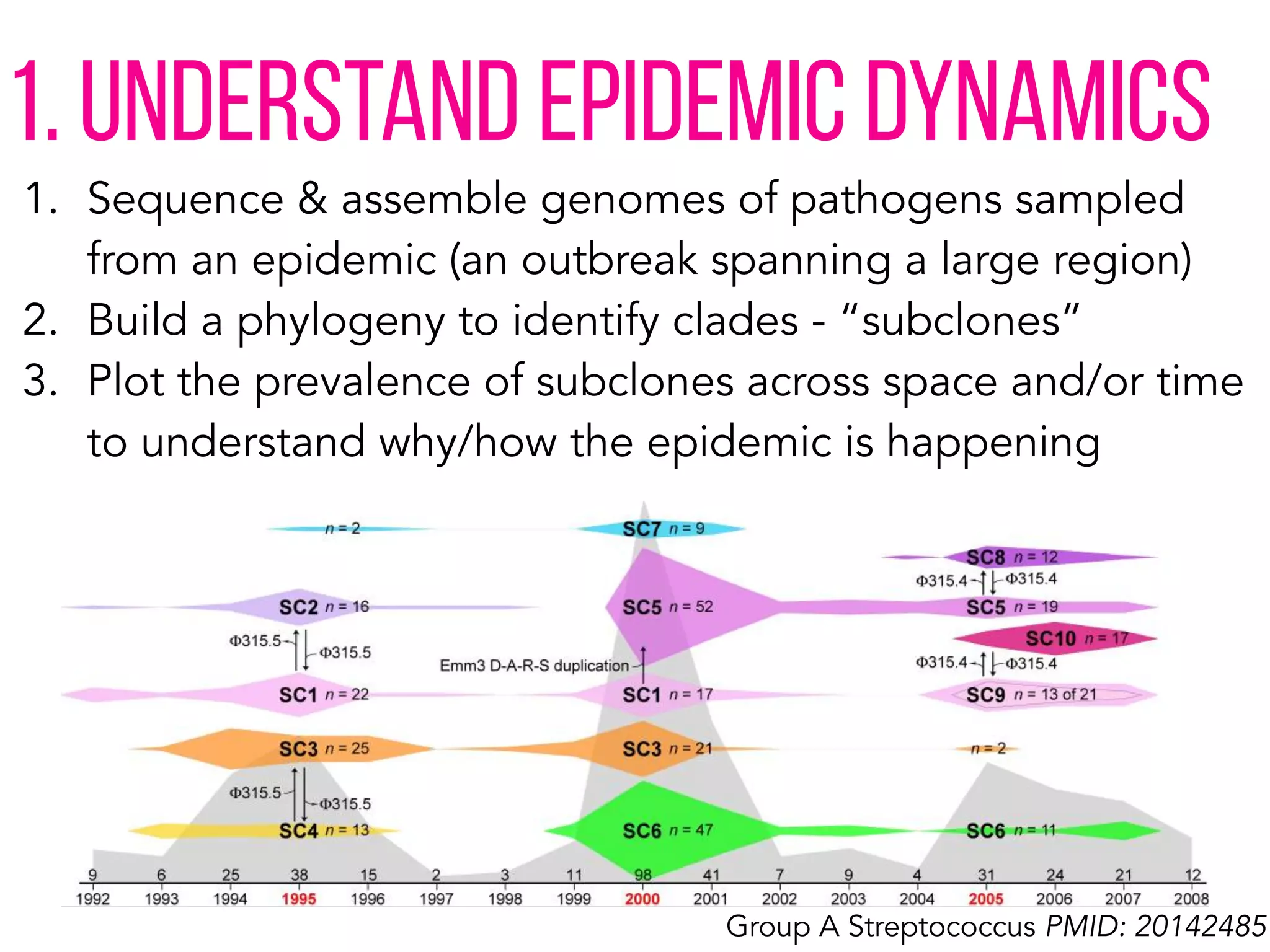
This document discusses how genomics is revolutionizing public health microbiology. It provides three examples:
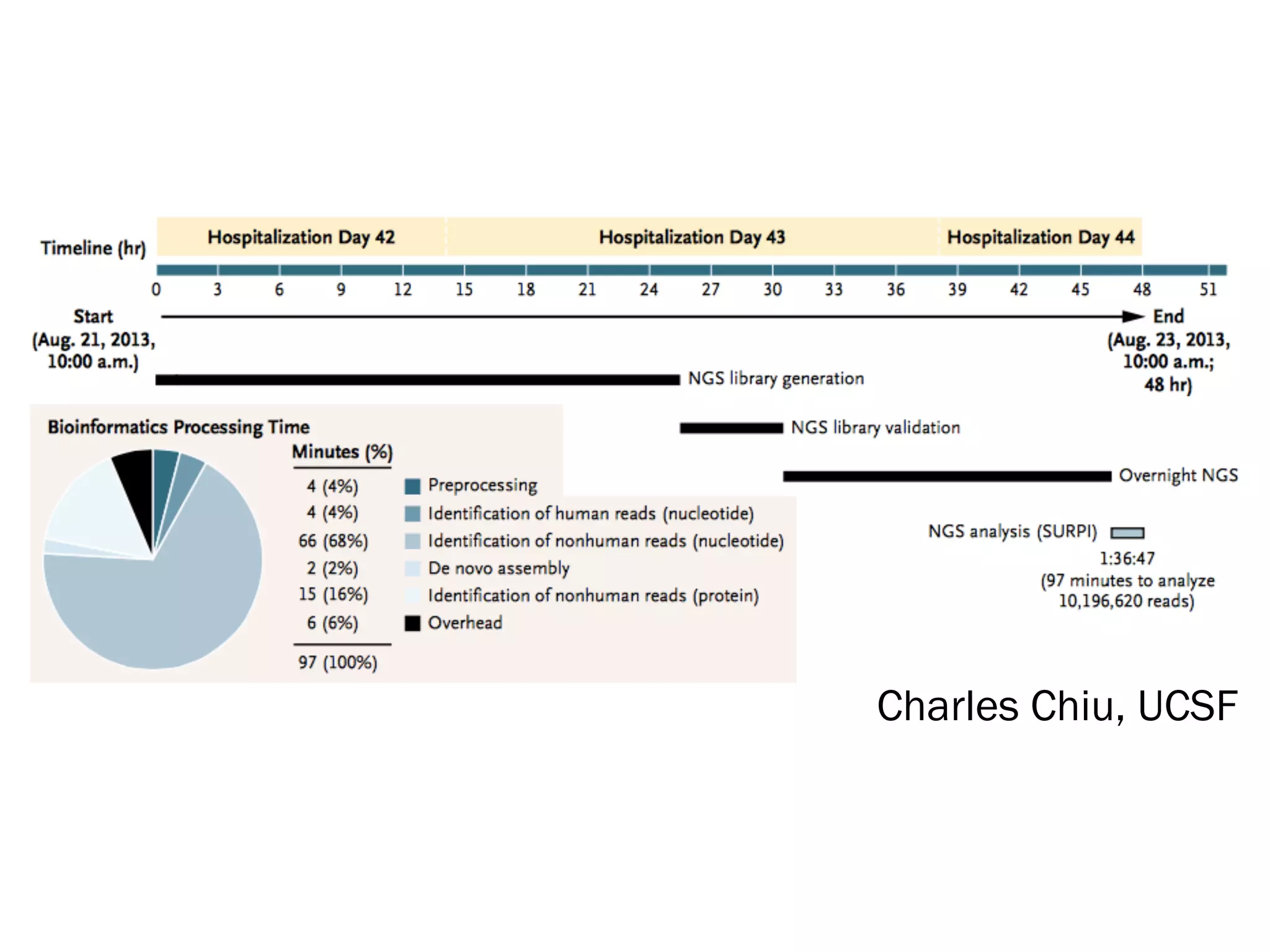
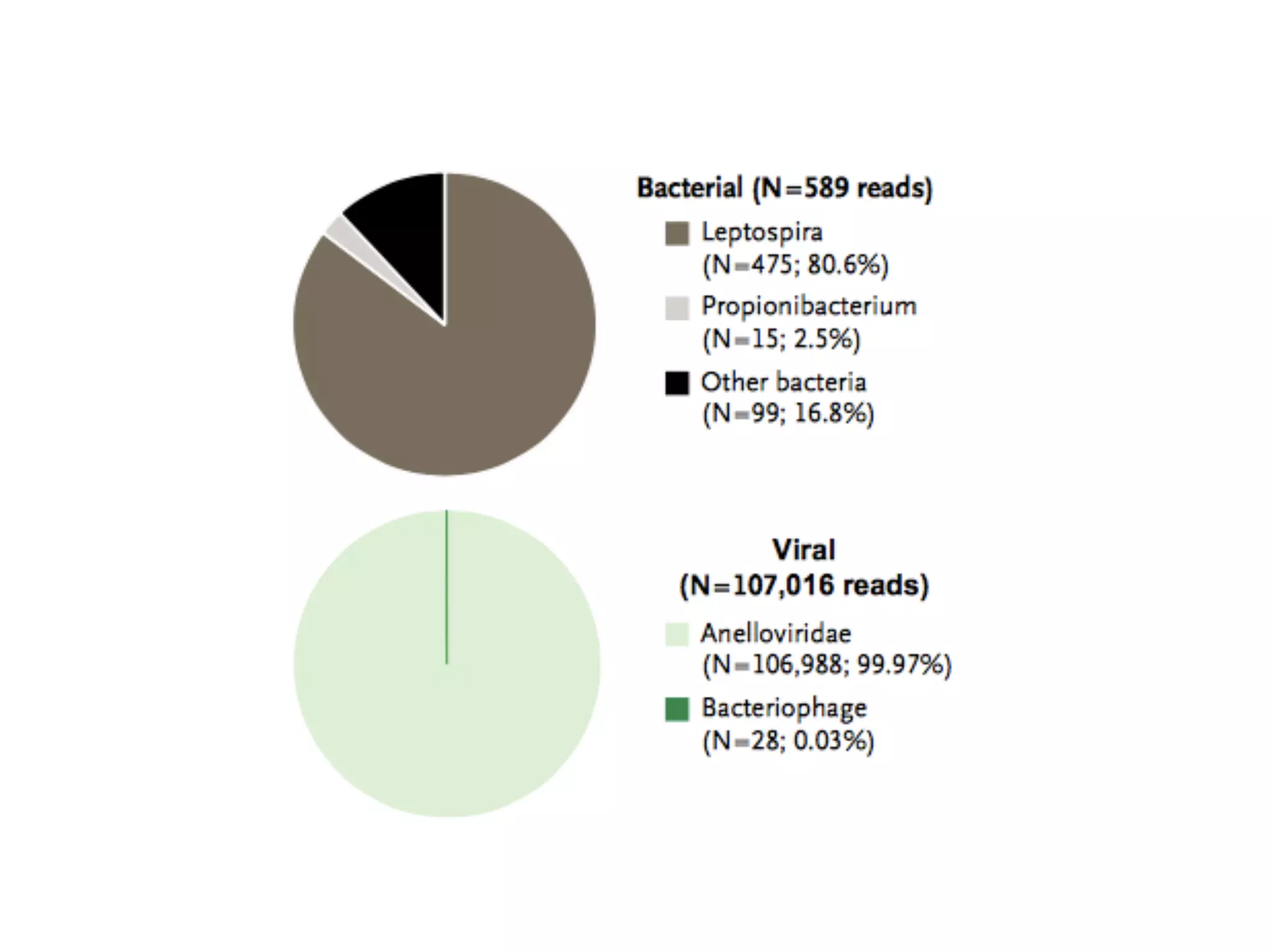
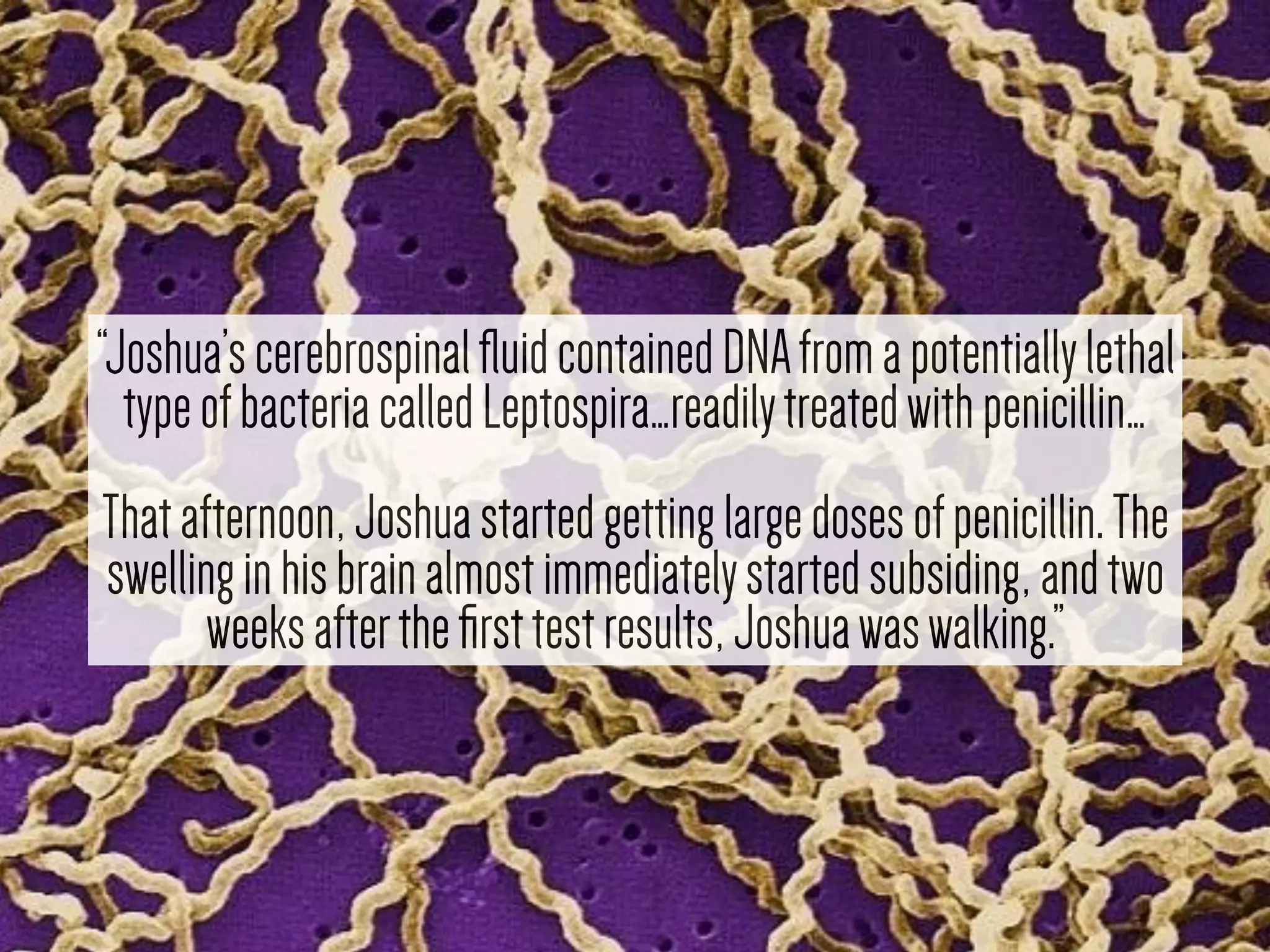
1) Rapid WGS-based diagnosis identified Leptospira bacteria in a patient's cerebrospinal fluid, allowing targeted penicillin treatment that resolved his illness within two weeks.
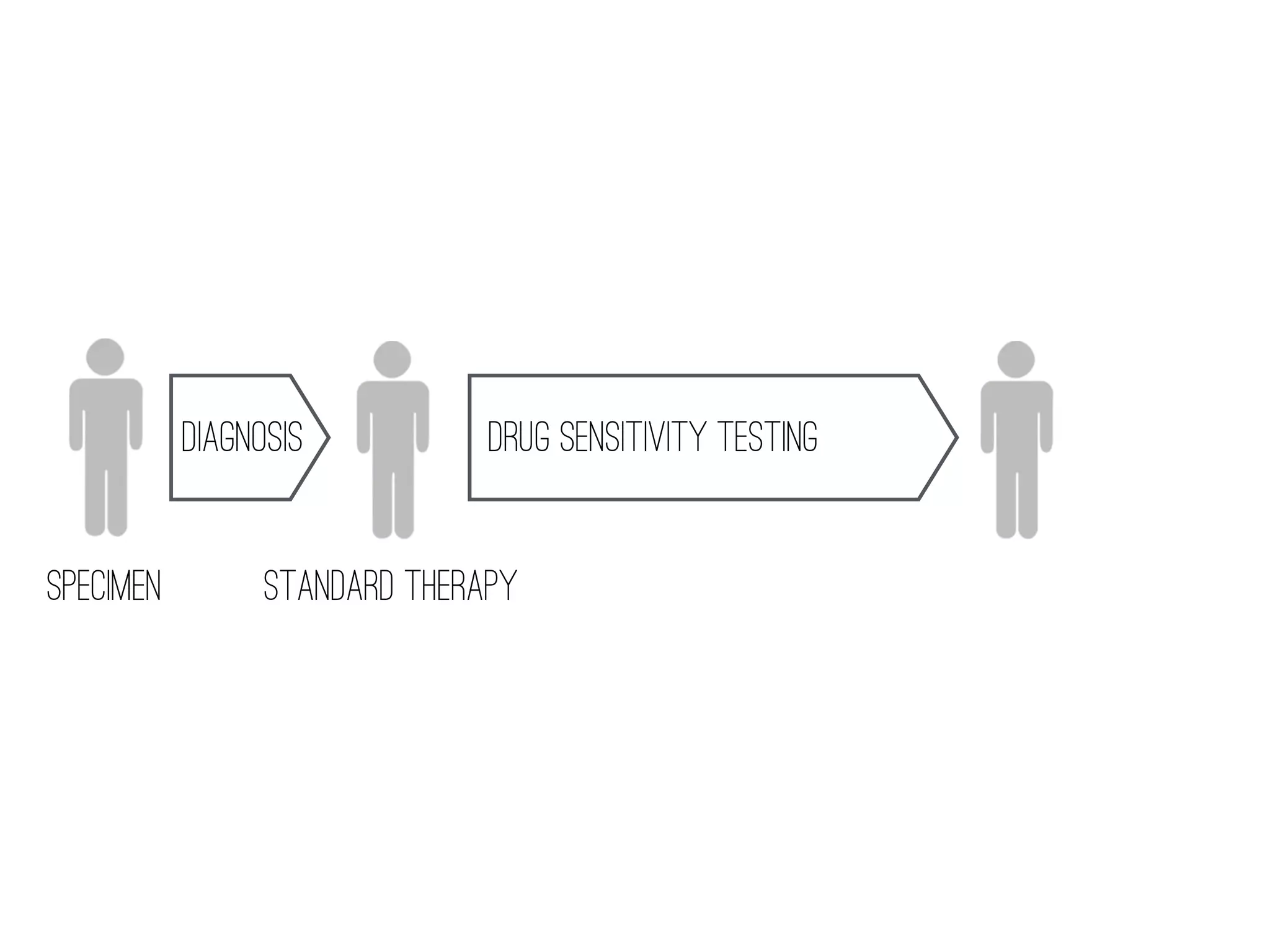
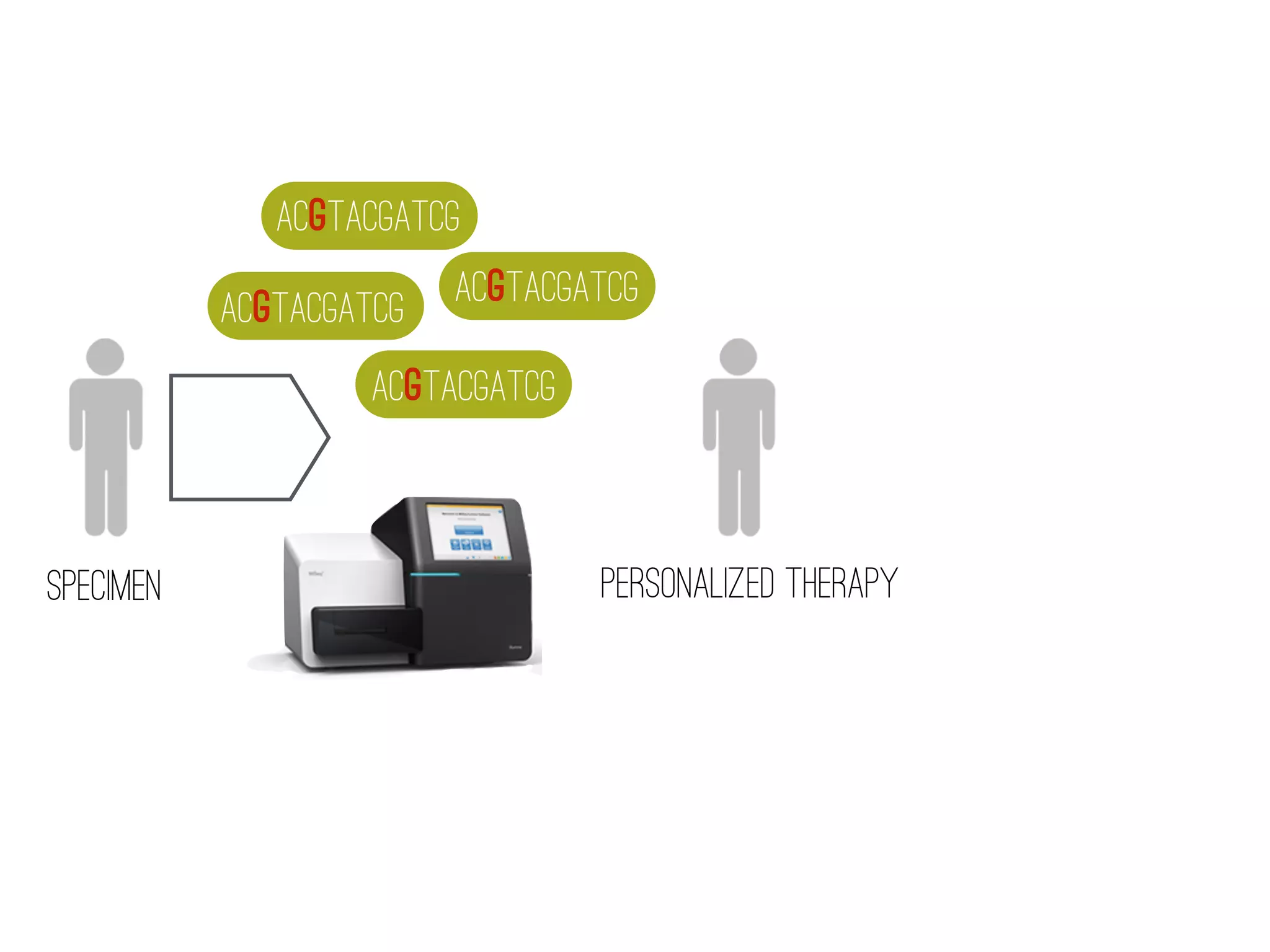
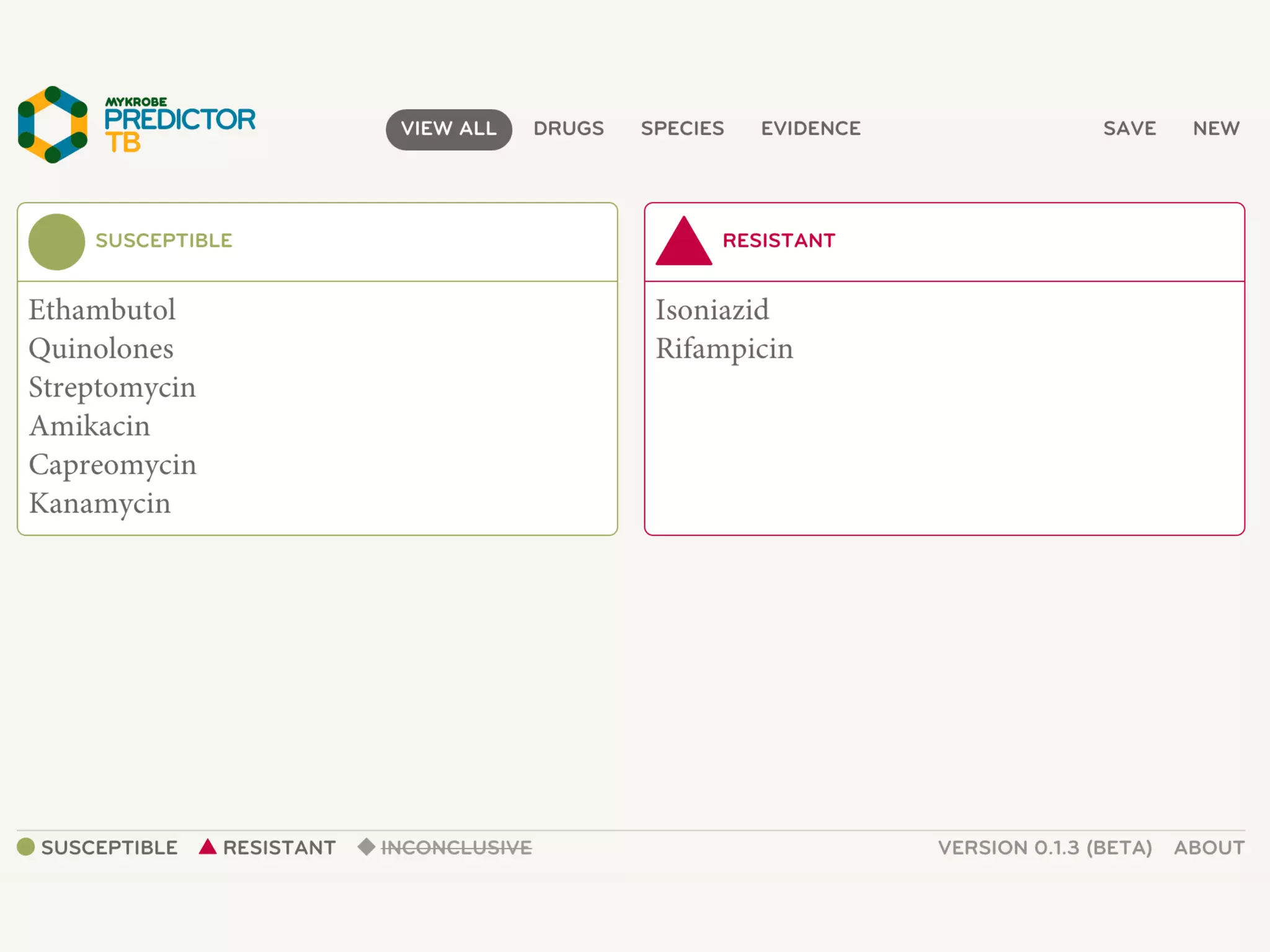
2) WGS-based drug sensitivity testing allows personalized therapy by predicting resistance from a pathogen's genome within 1 day, compared to weeks for conventional methods.
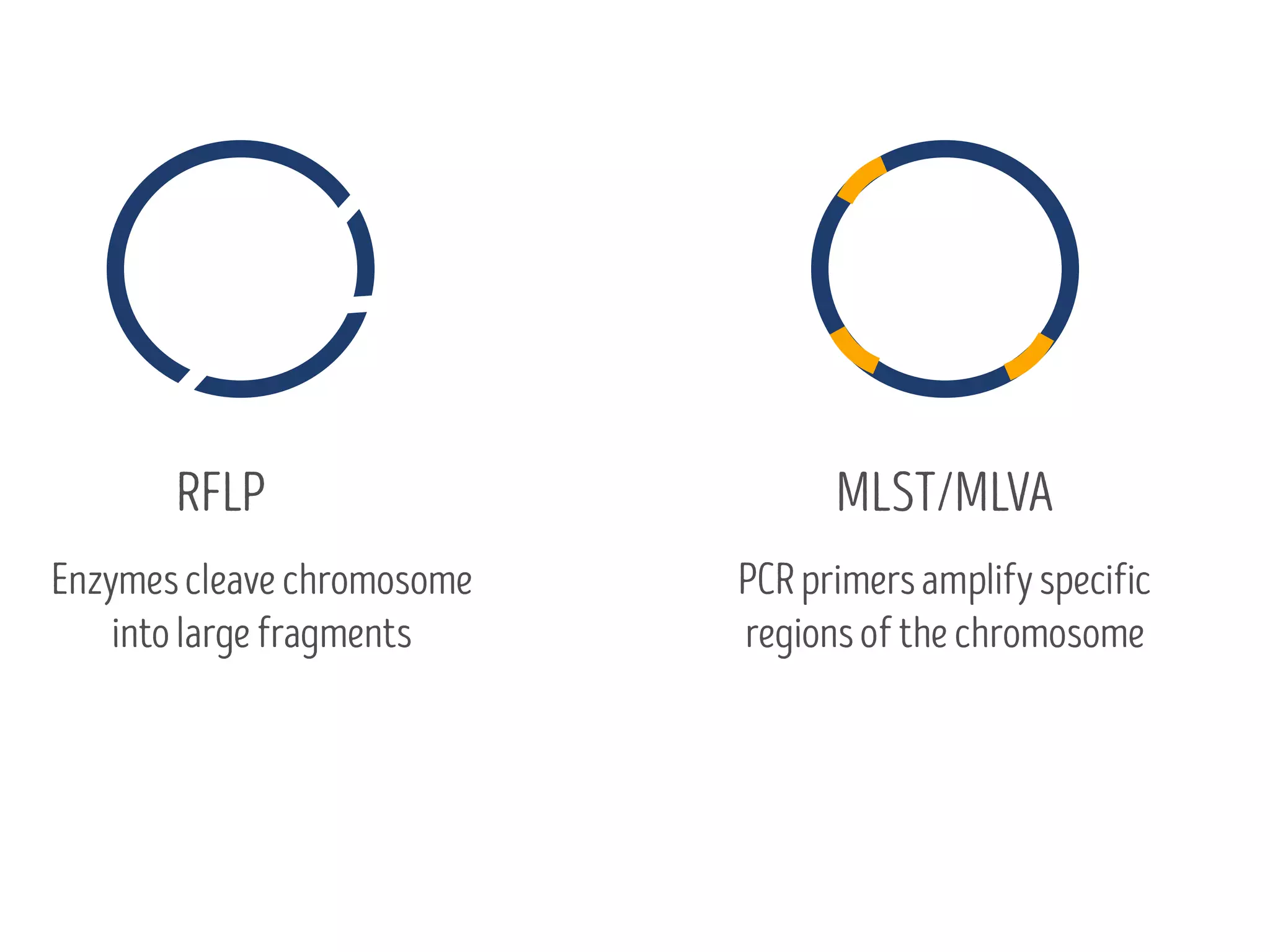
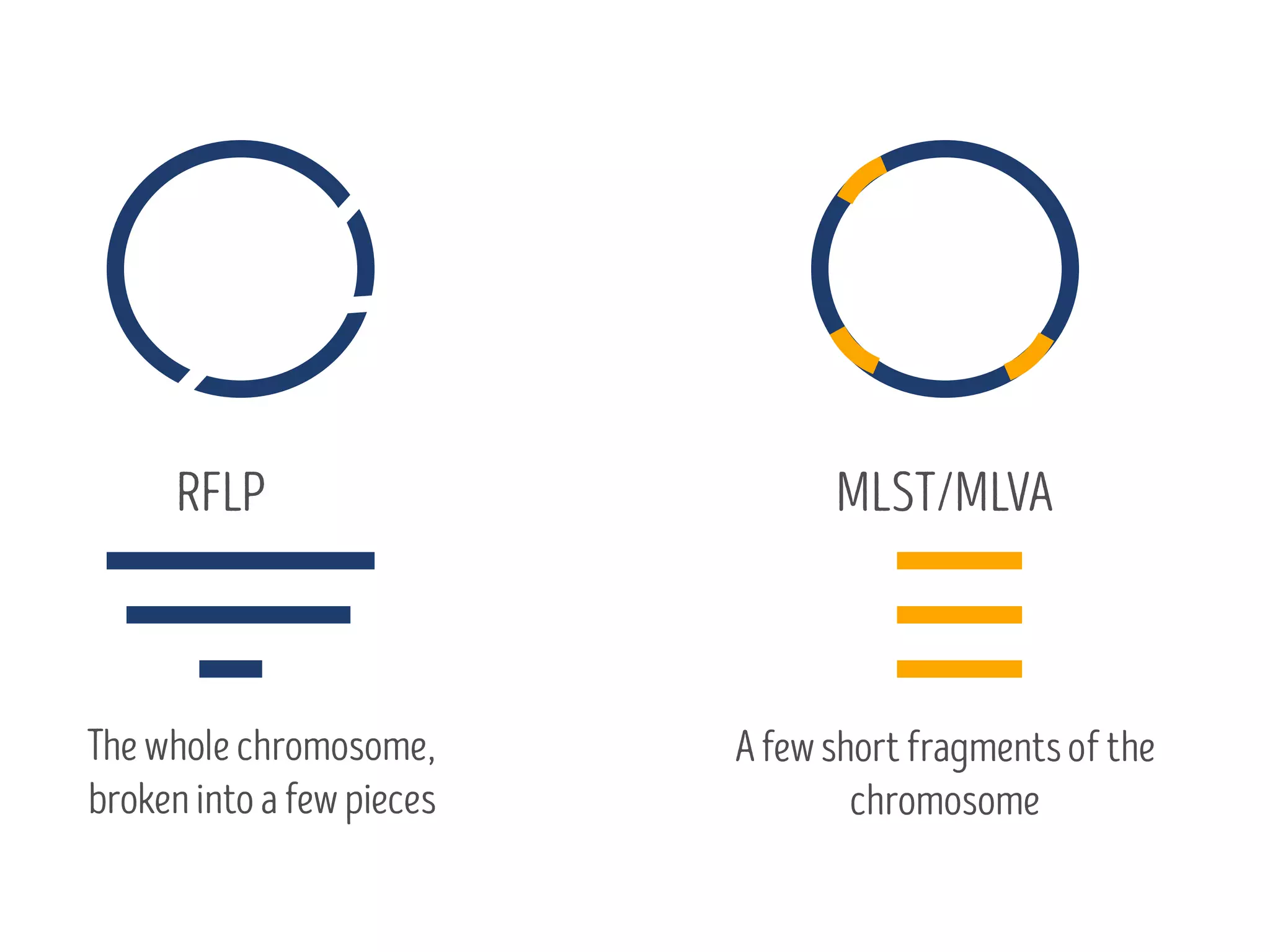
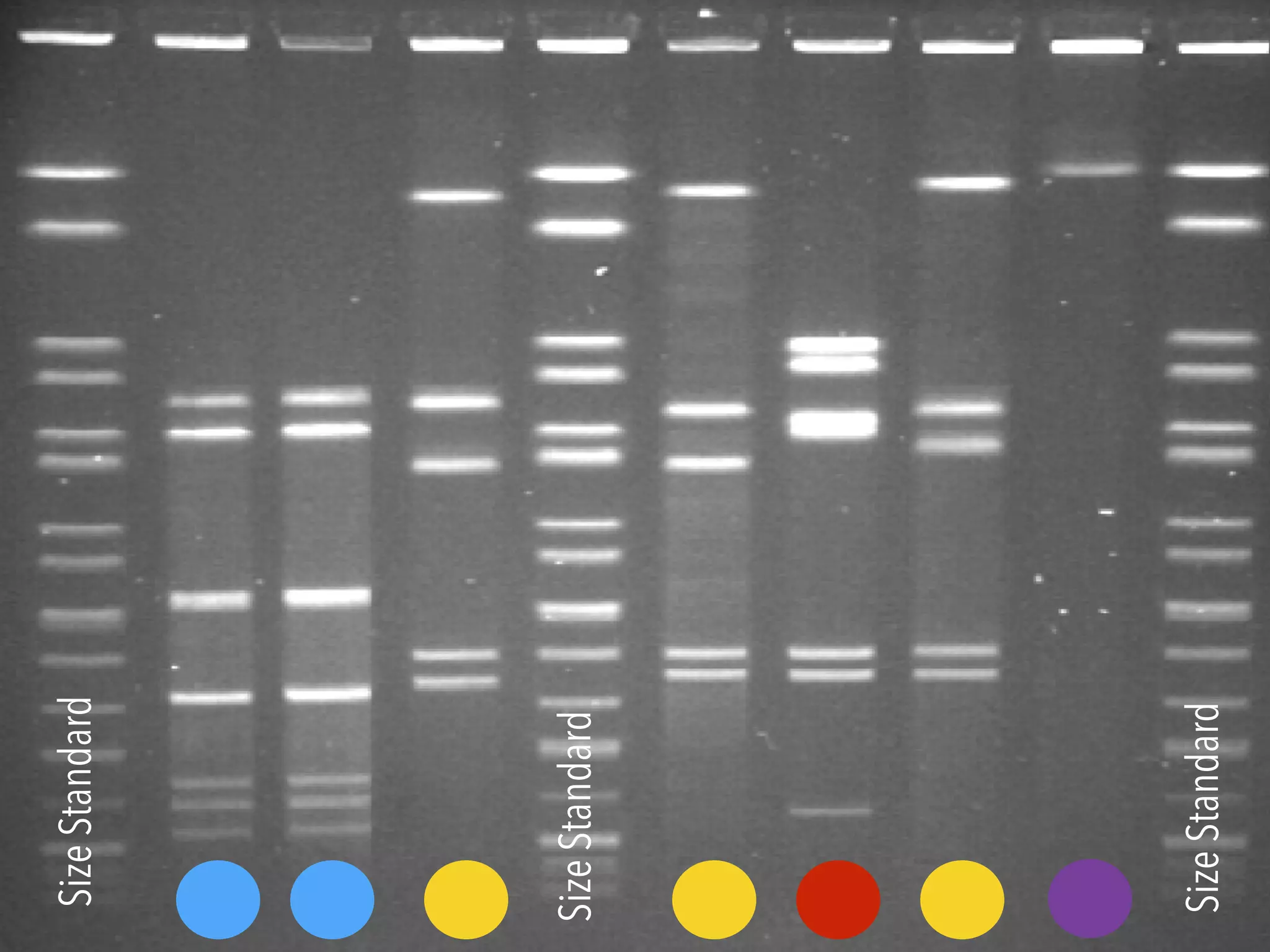
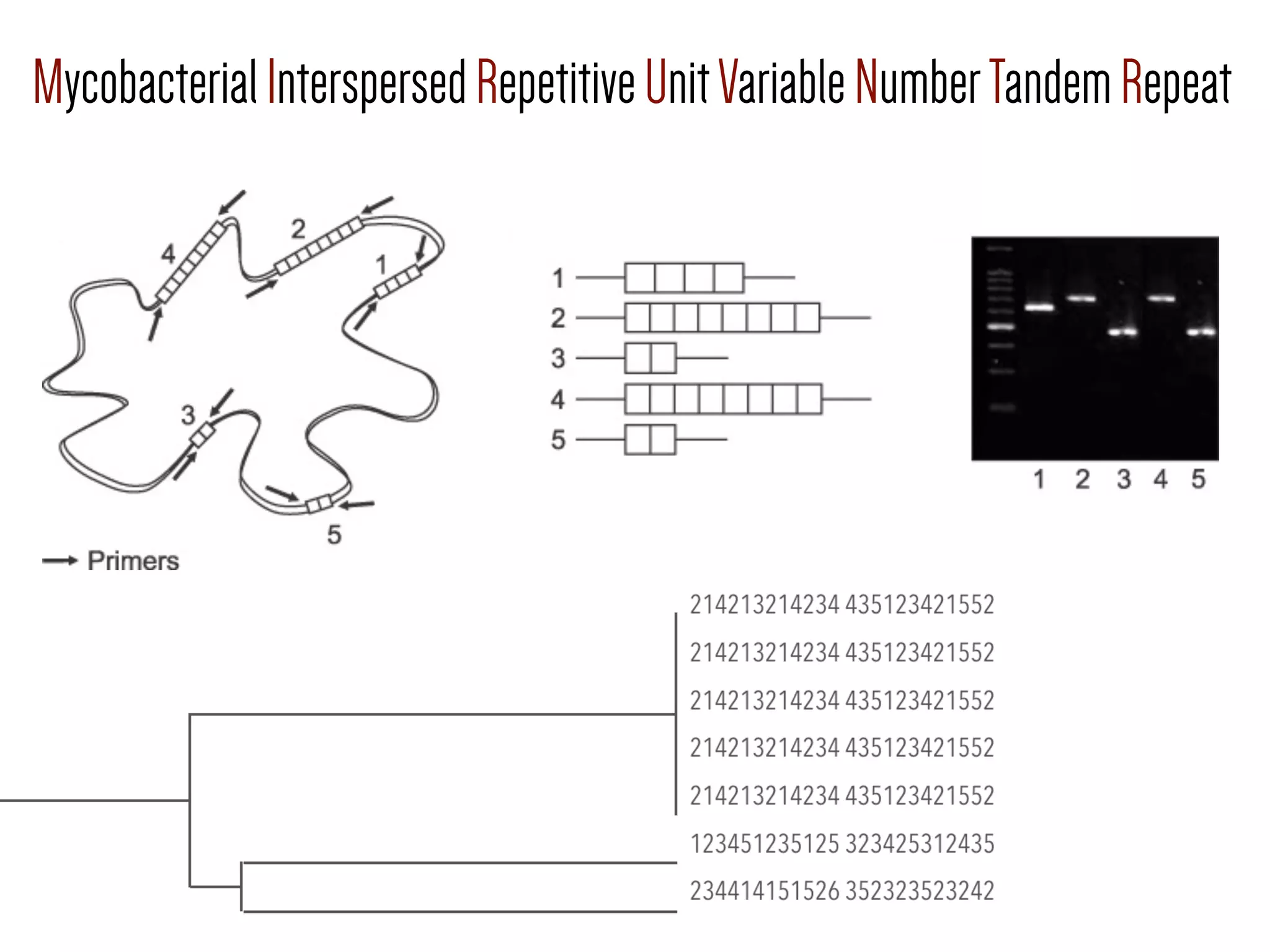
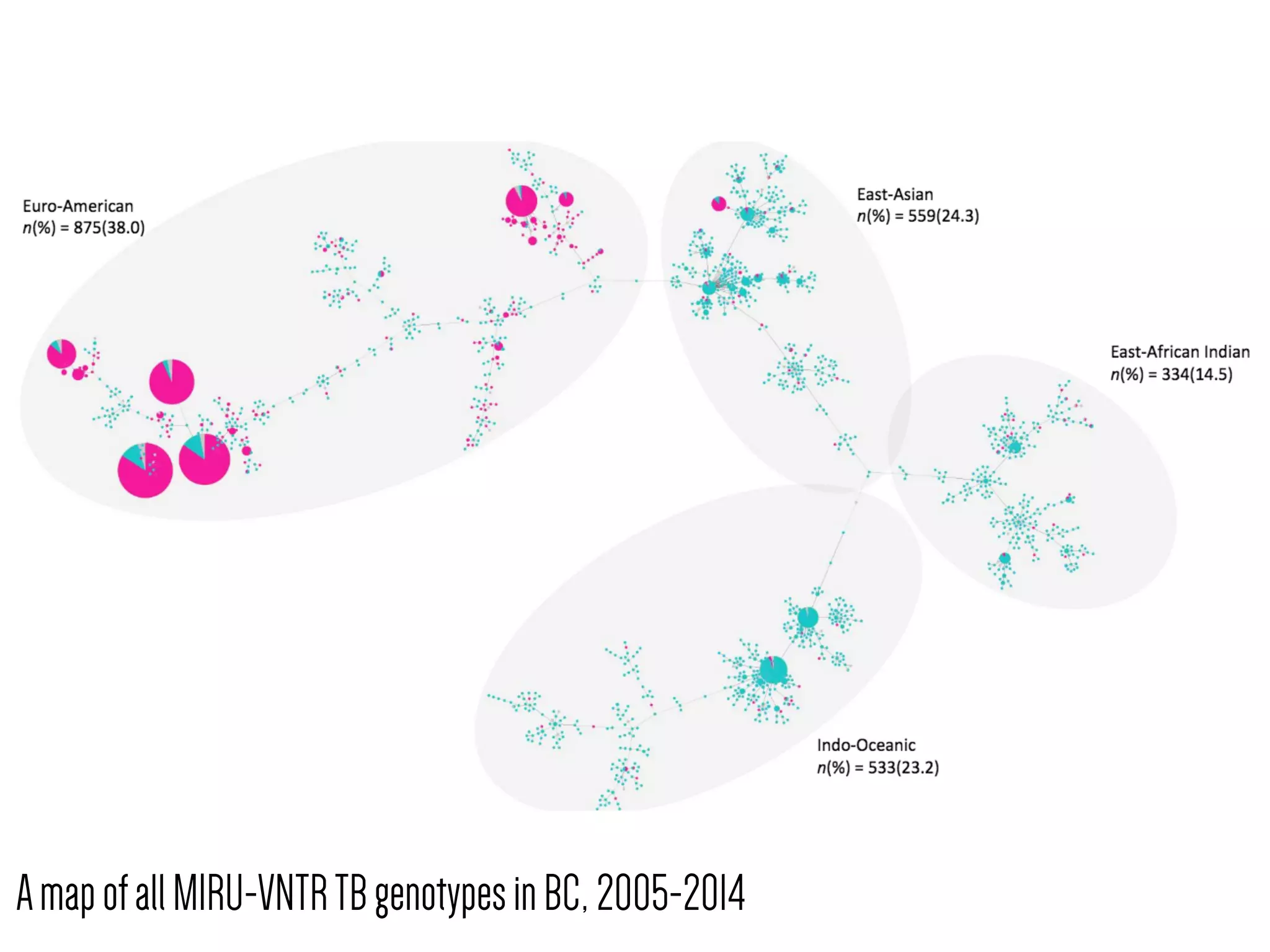
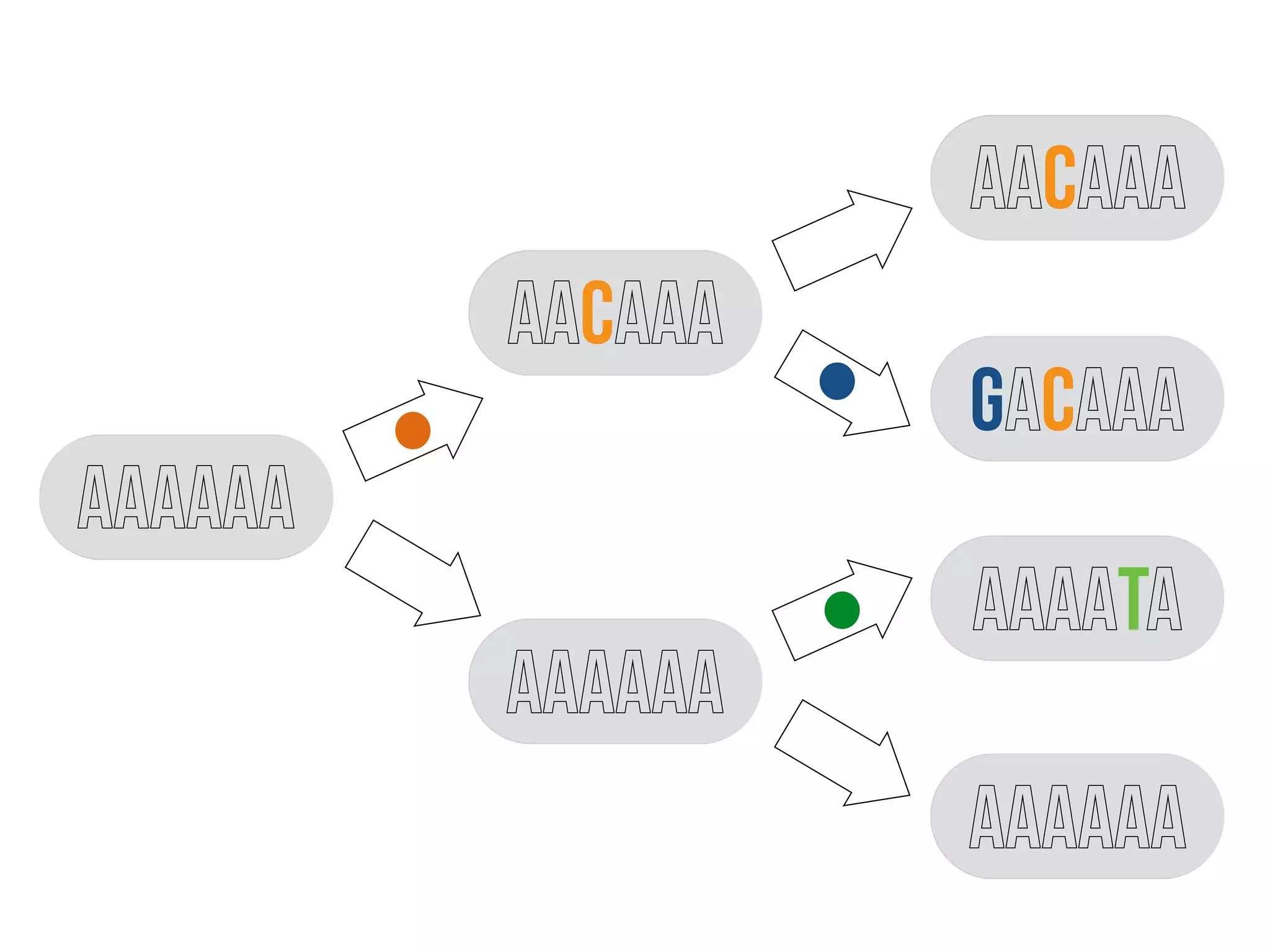
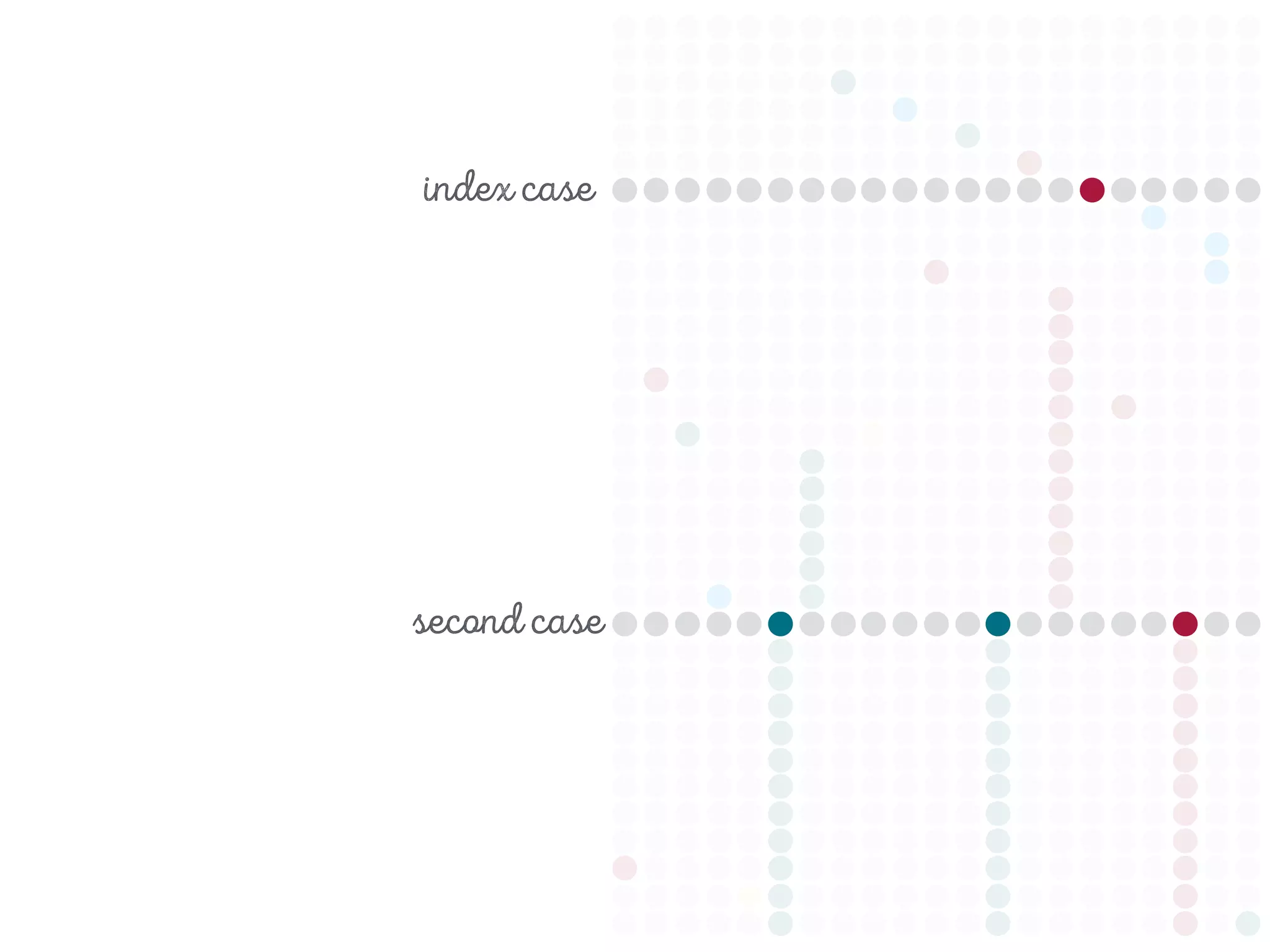
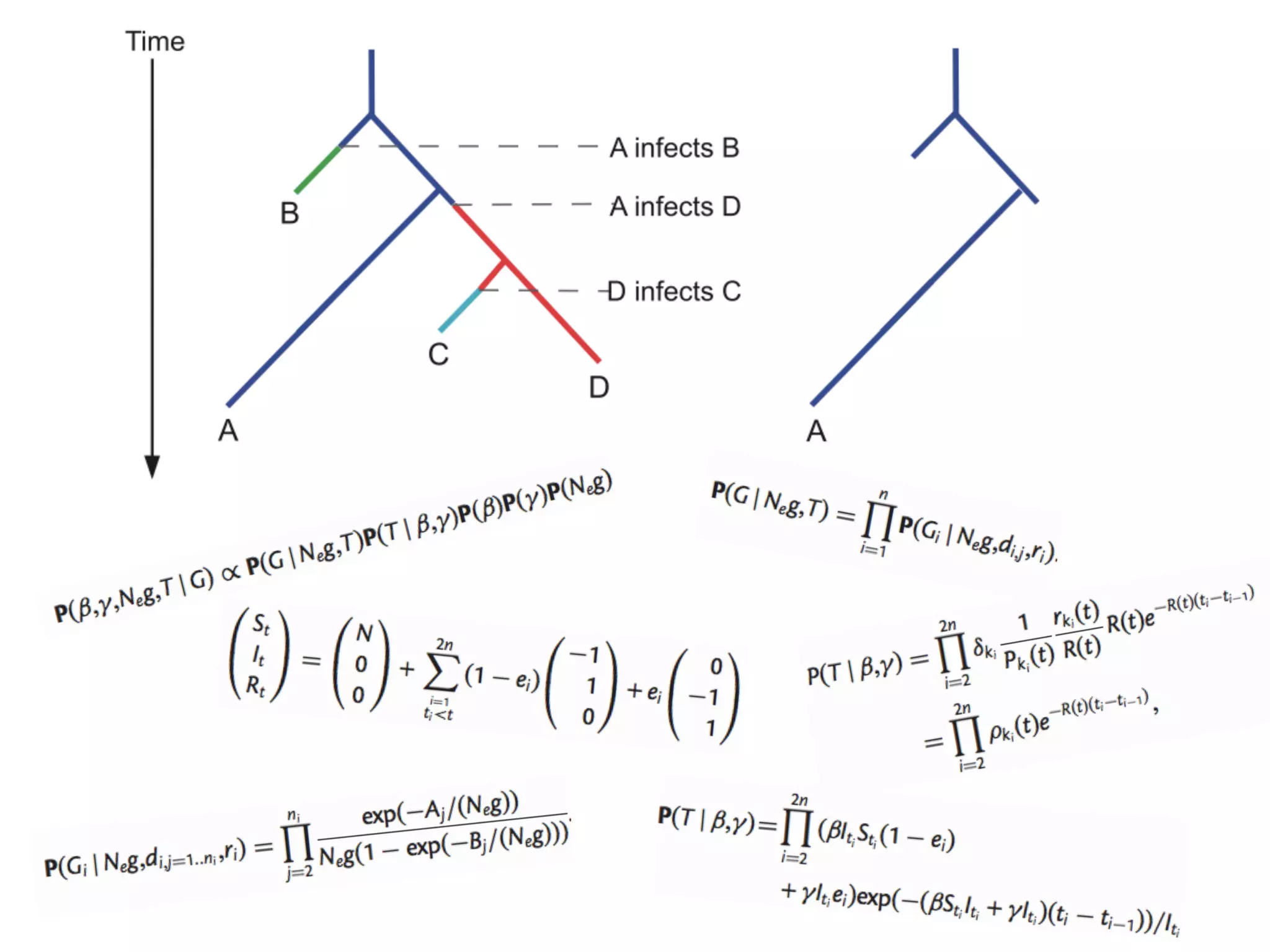
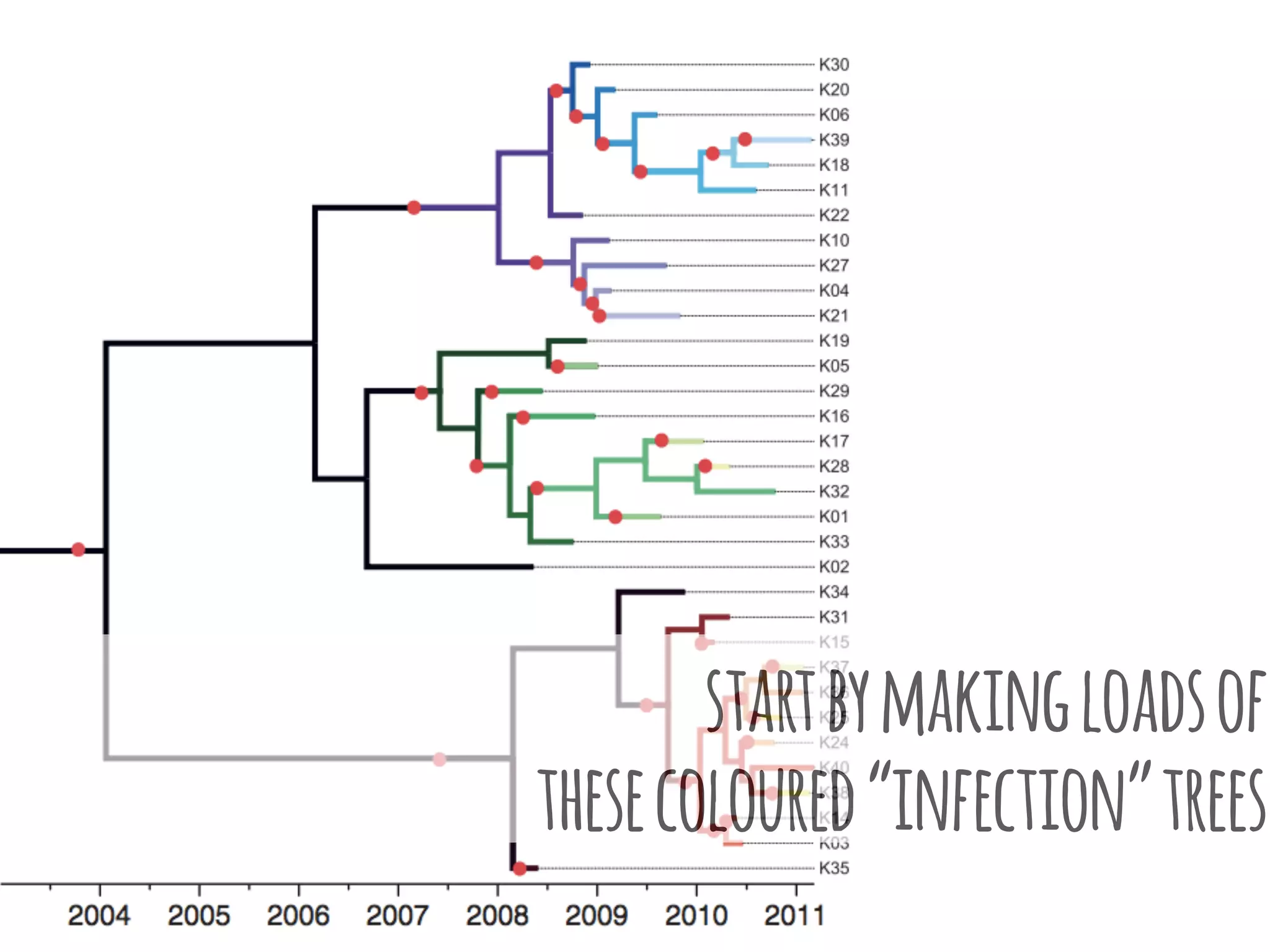
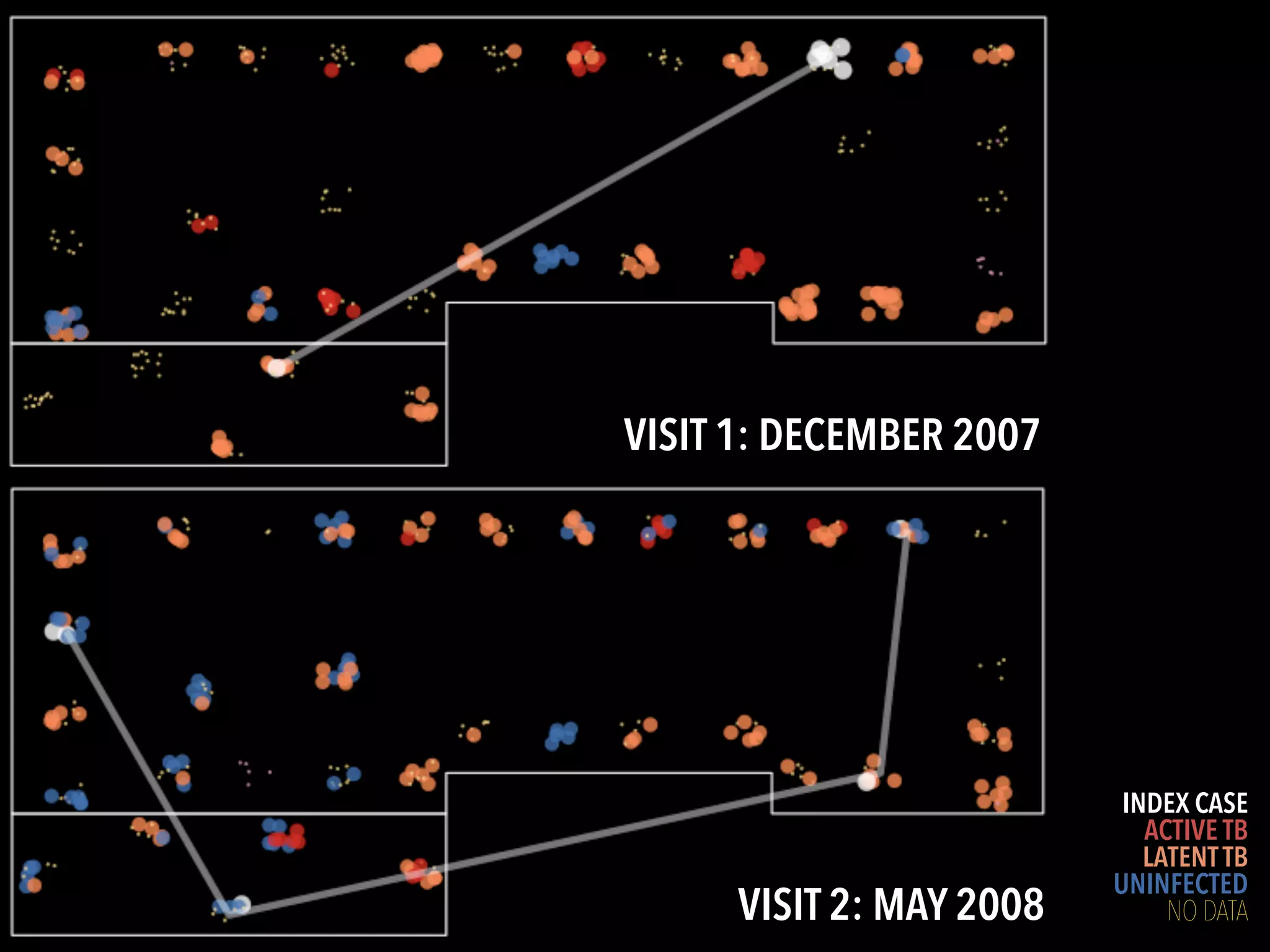
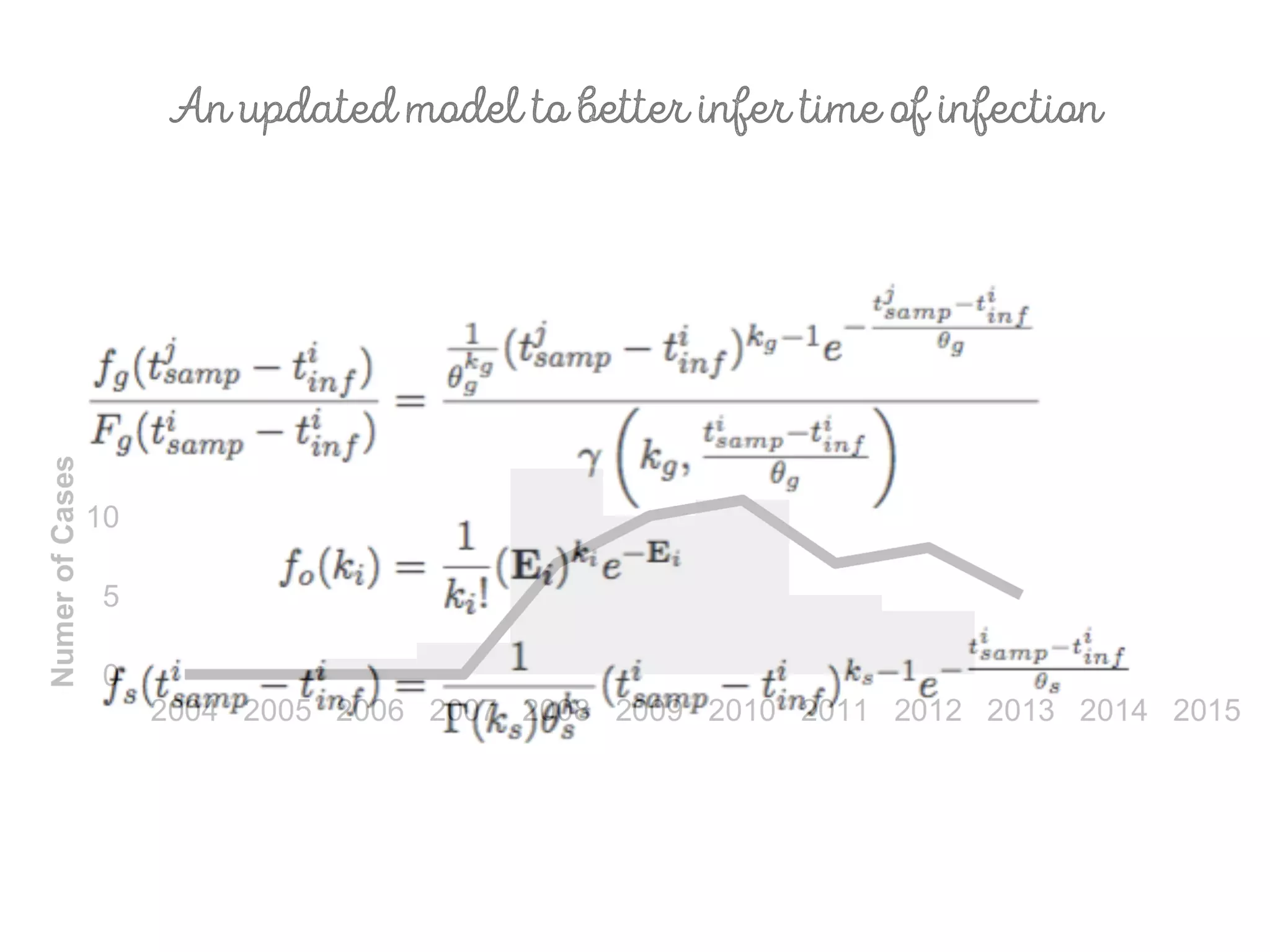
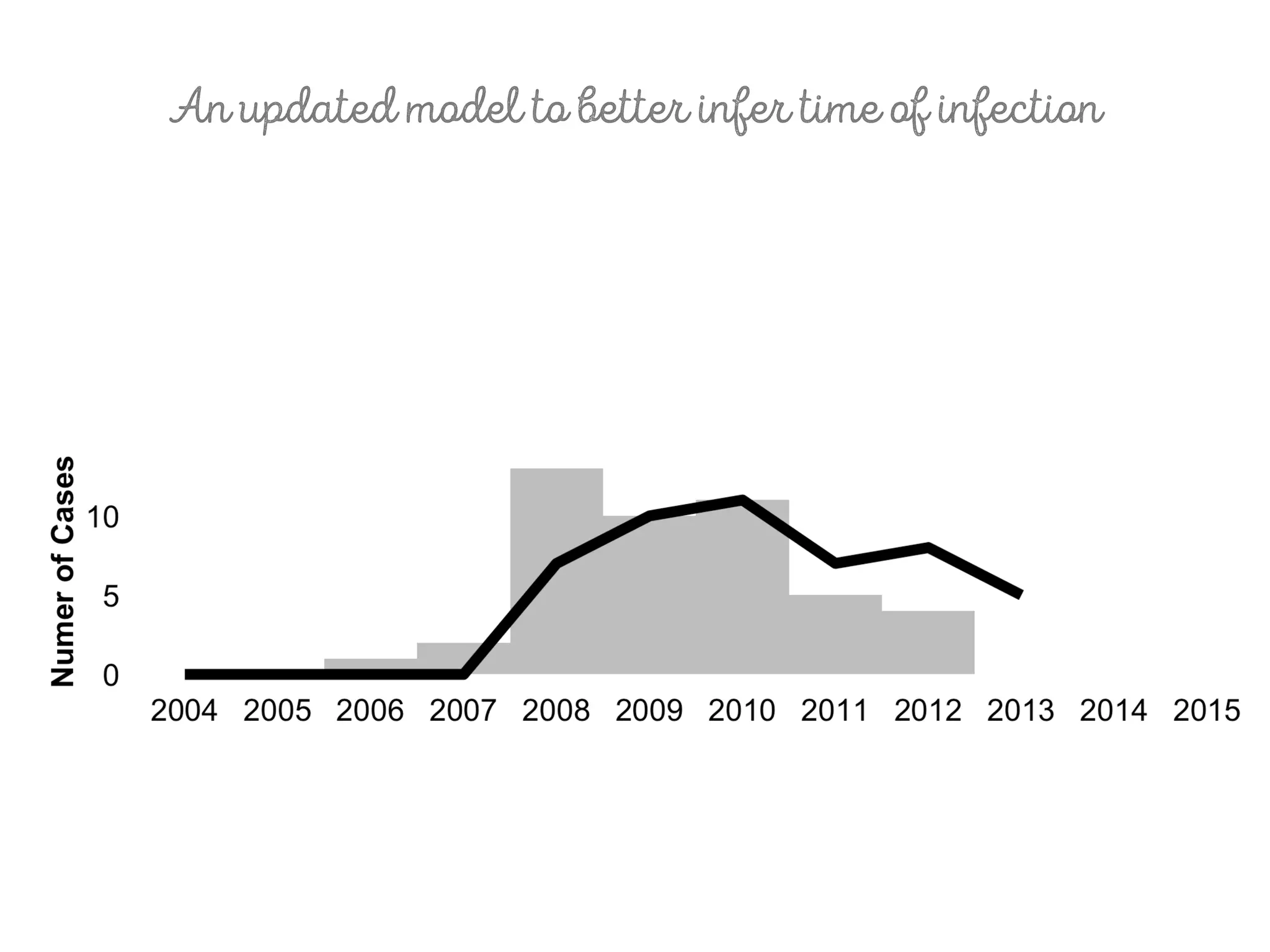
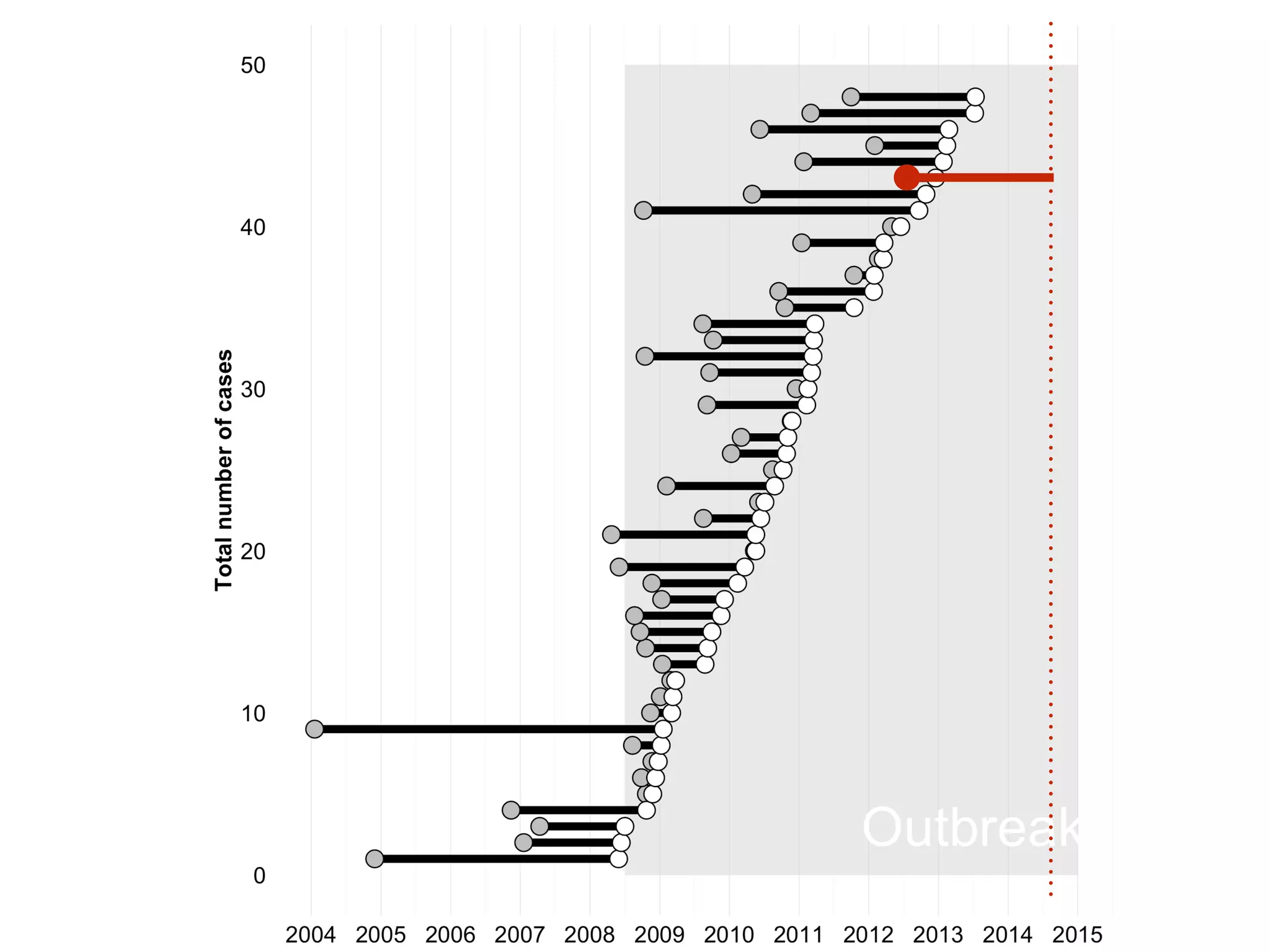
3) Genomic epidemiology tracks person-to-person disease spread by comparing whole genomes from outbreak isolates. This approach helped determine the key locations and time period fueling a TB outbreak in BC, and later demonstrated transmission had ended.