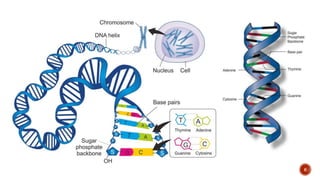
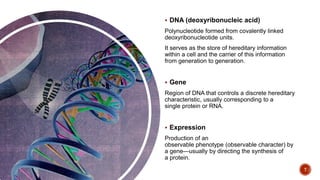
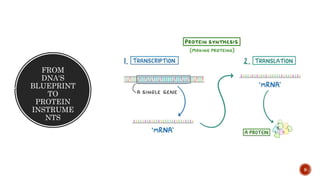
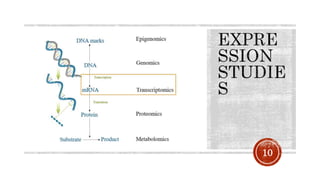
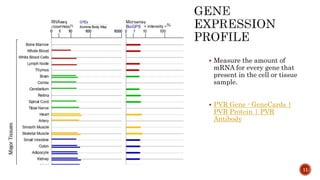
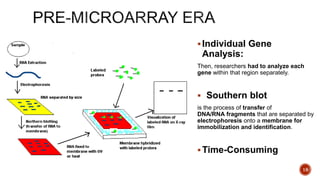
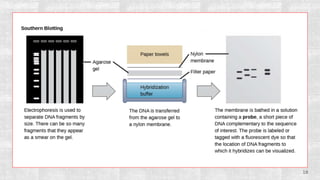
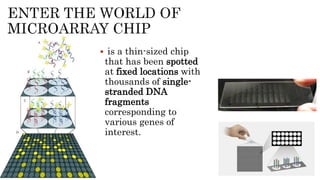
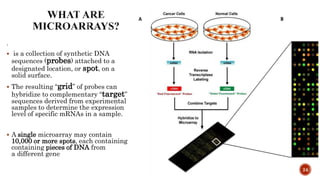
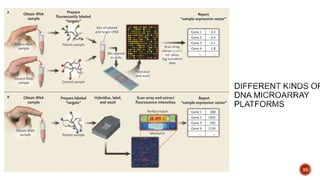
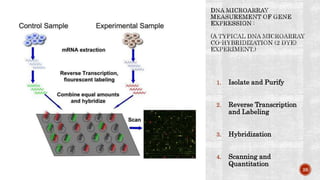
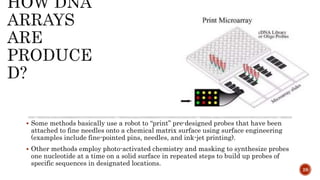
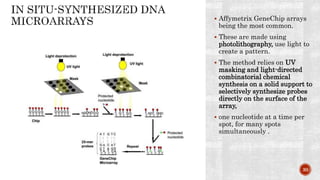
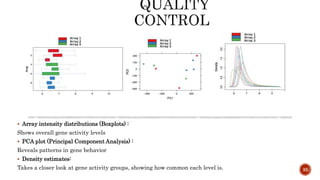
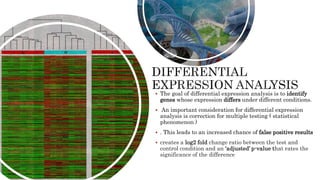
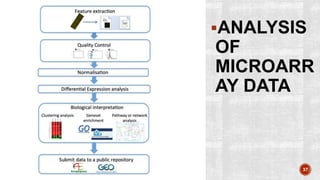
The document discusses the role of microarrays in decoding genetic information, notably focusing on the historical contributions of Rosalind Franklin and the Nobel award winners for discovering DNA's structure. It outlines the technical aspects of microarrays for analyzing gene expression, their advantages in high-throughput screening, and challenges such as their limitations in detecting unknown genes. Furthermore, it highlights ongoing genomic research projects like the Qatar Genome Program, aimed at advancing personalized medicine and understanding genetic conditions in specific populations.