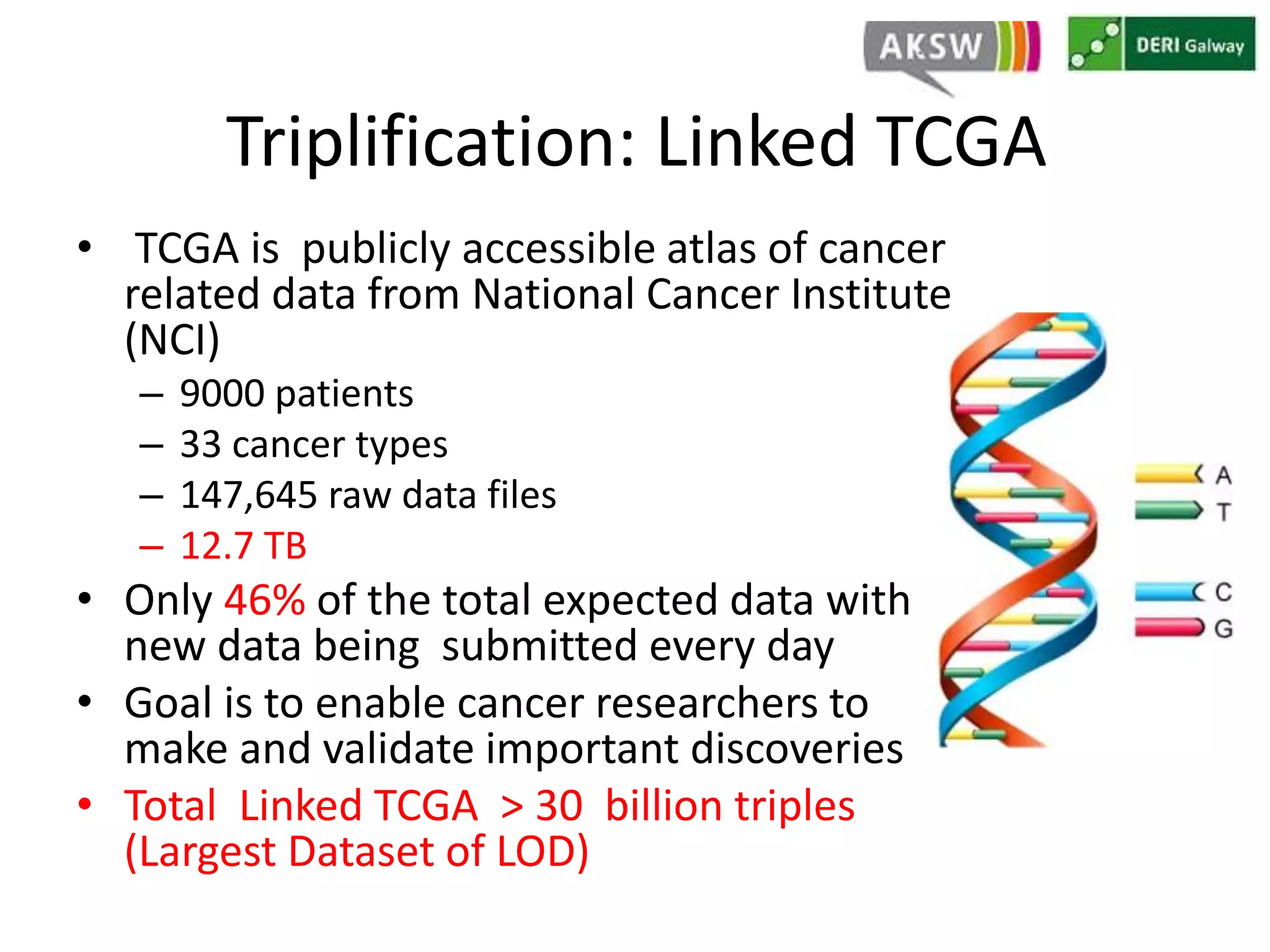
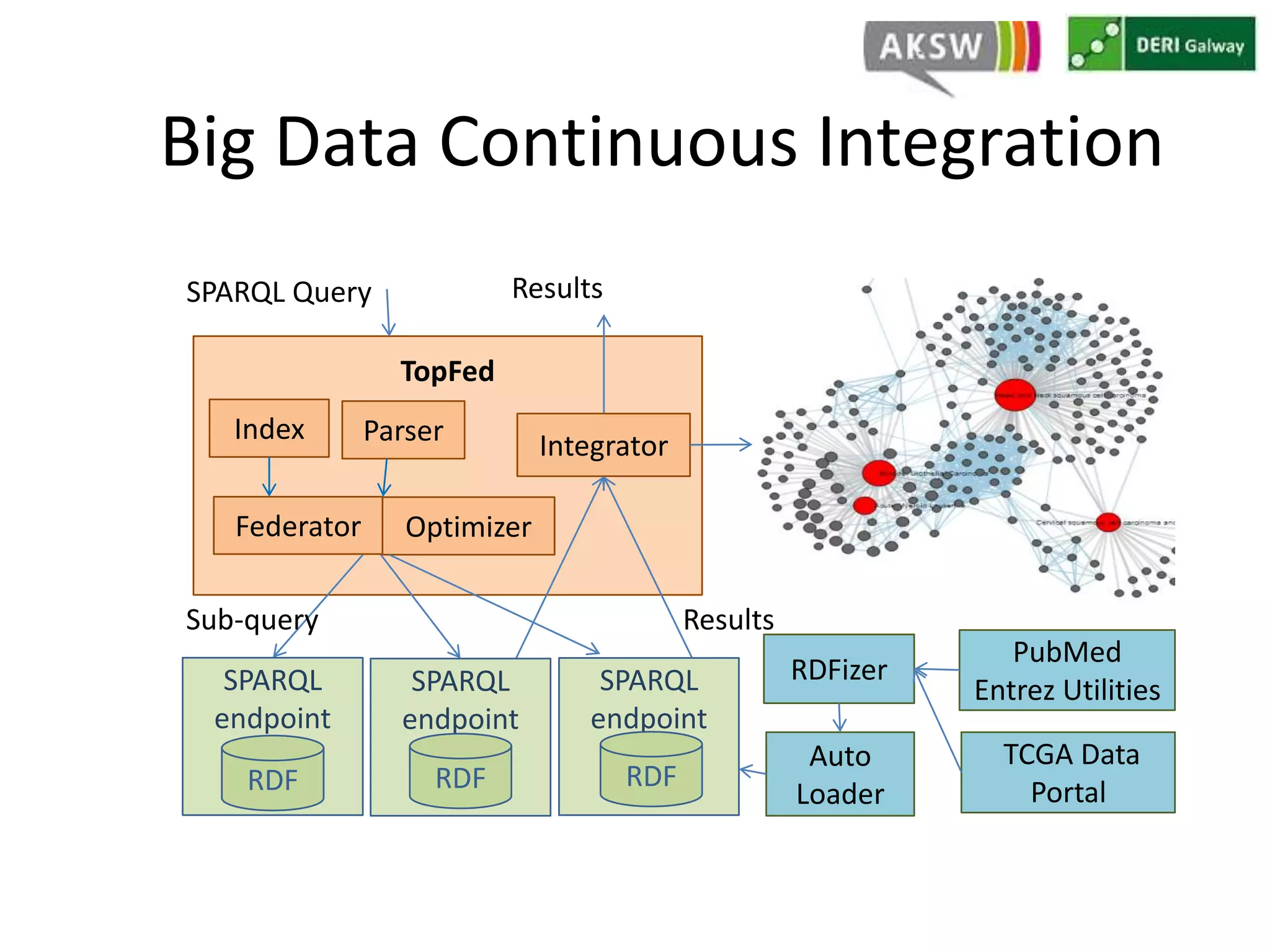
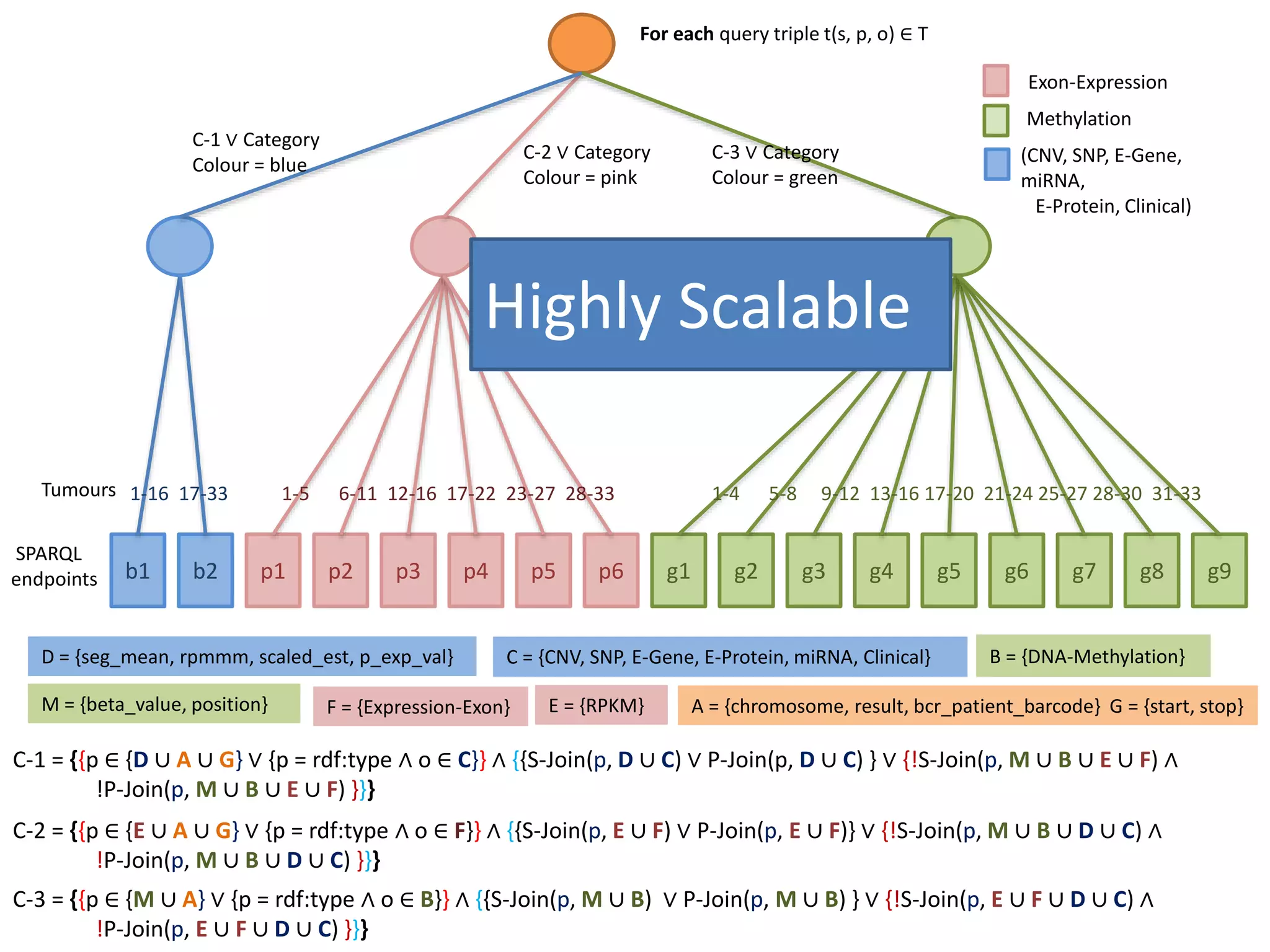
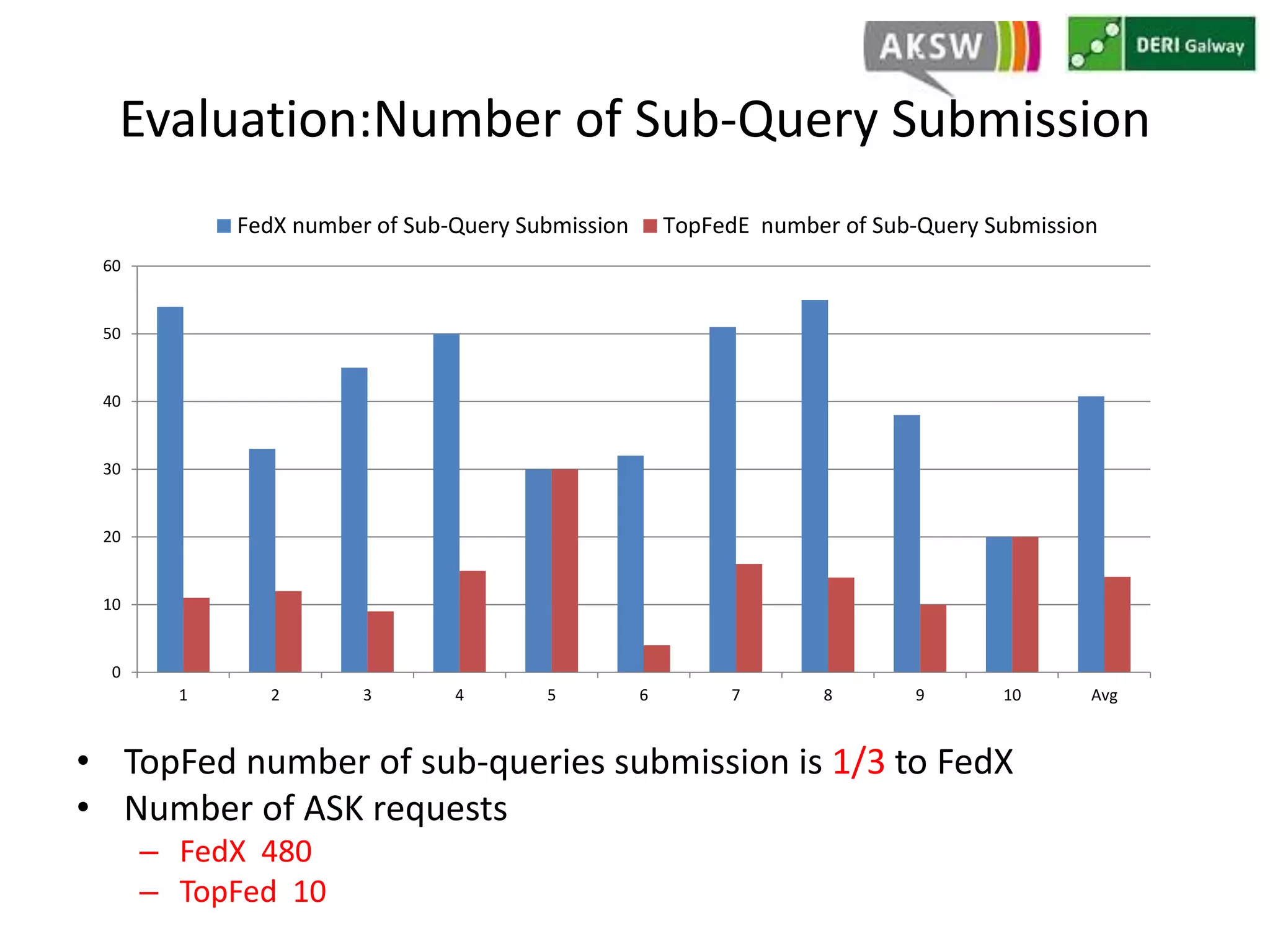
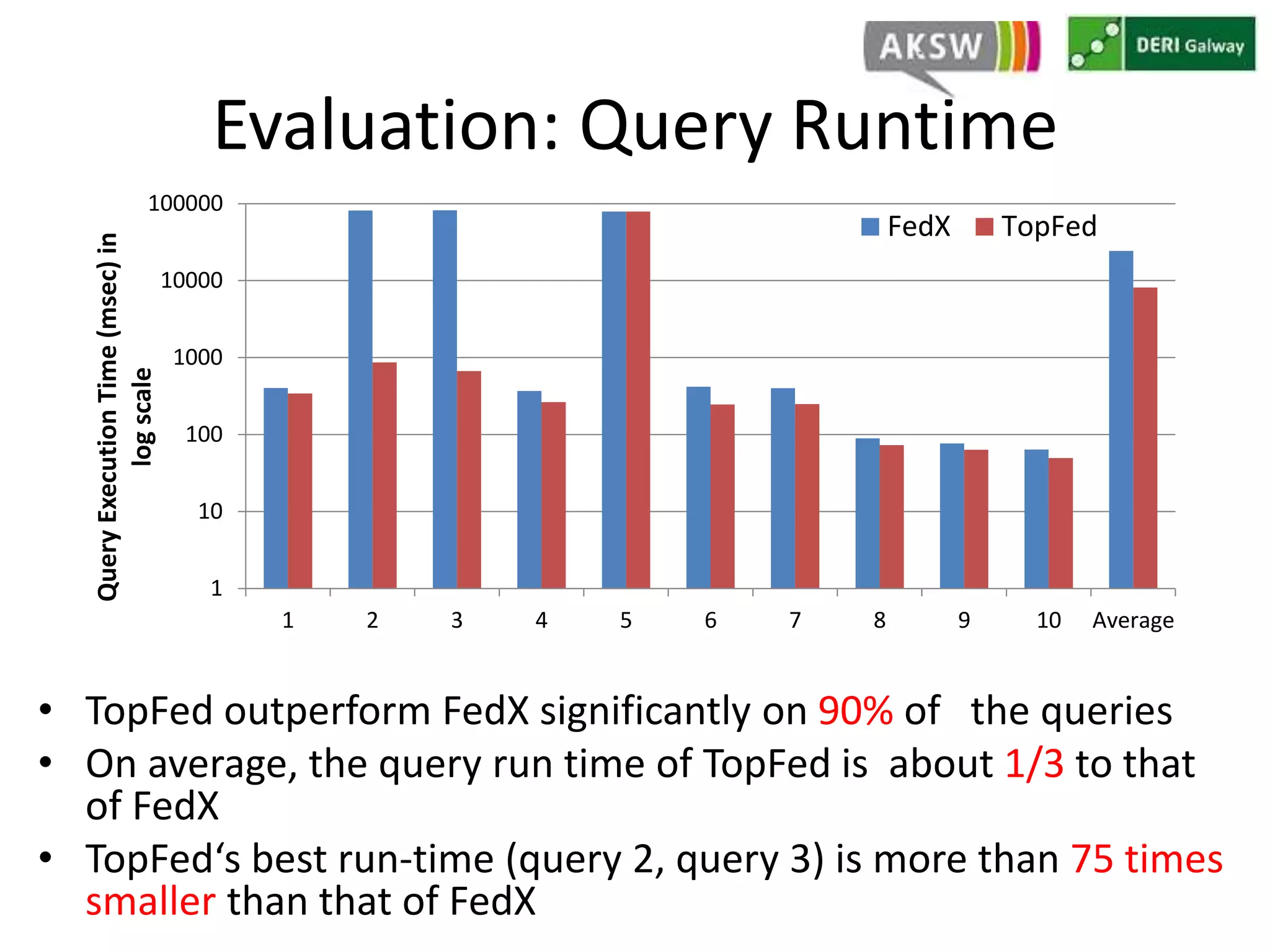
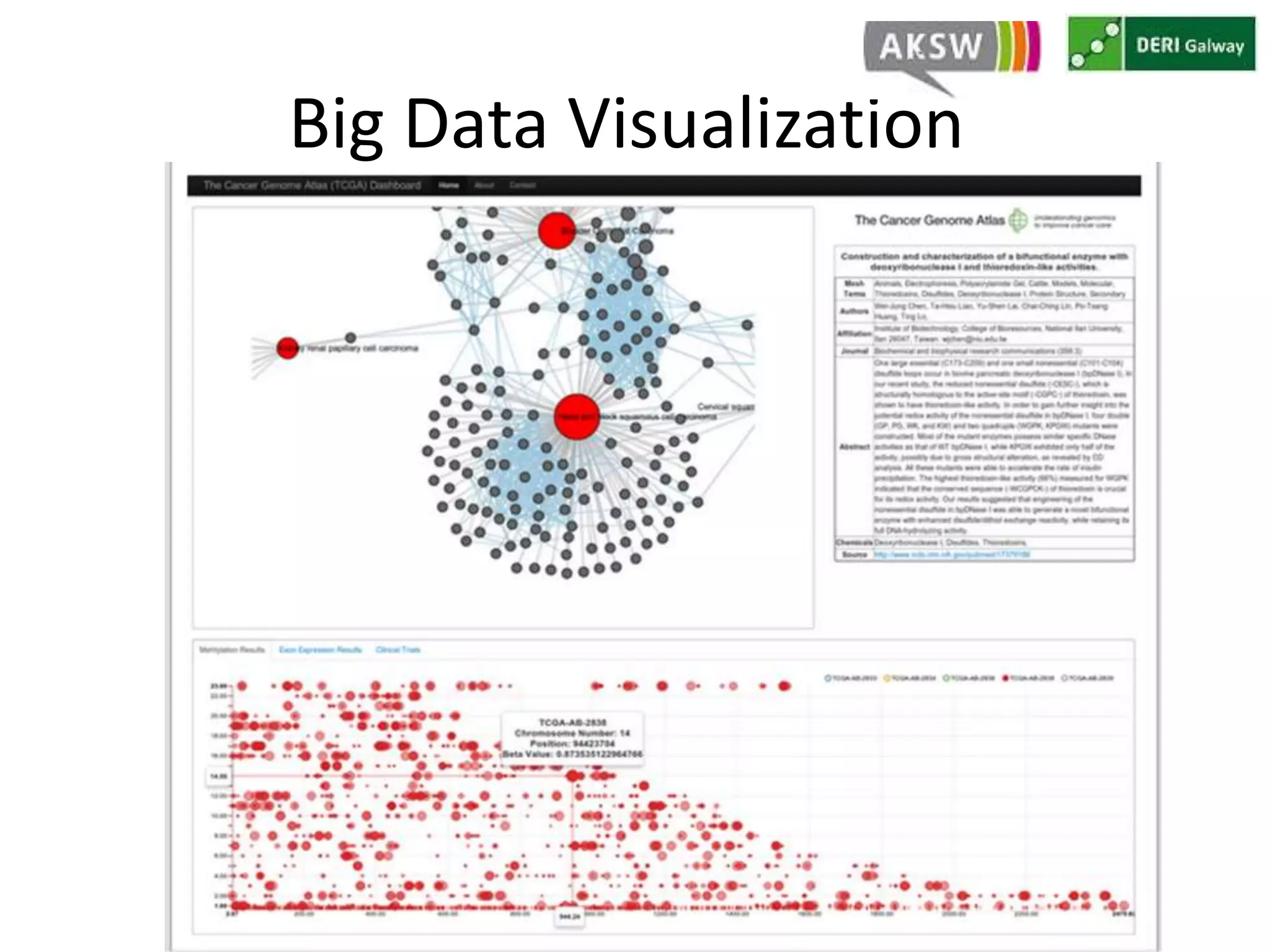
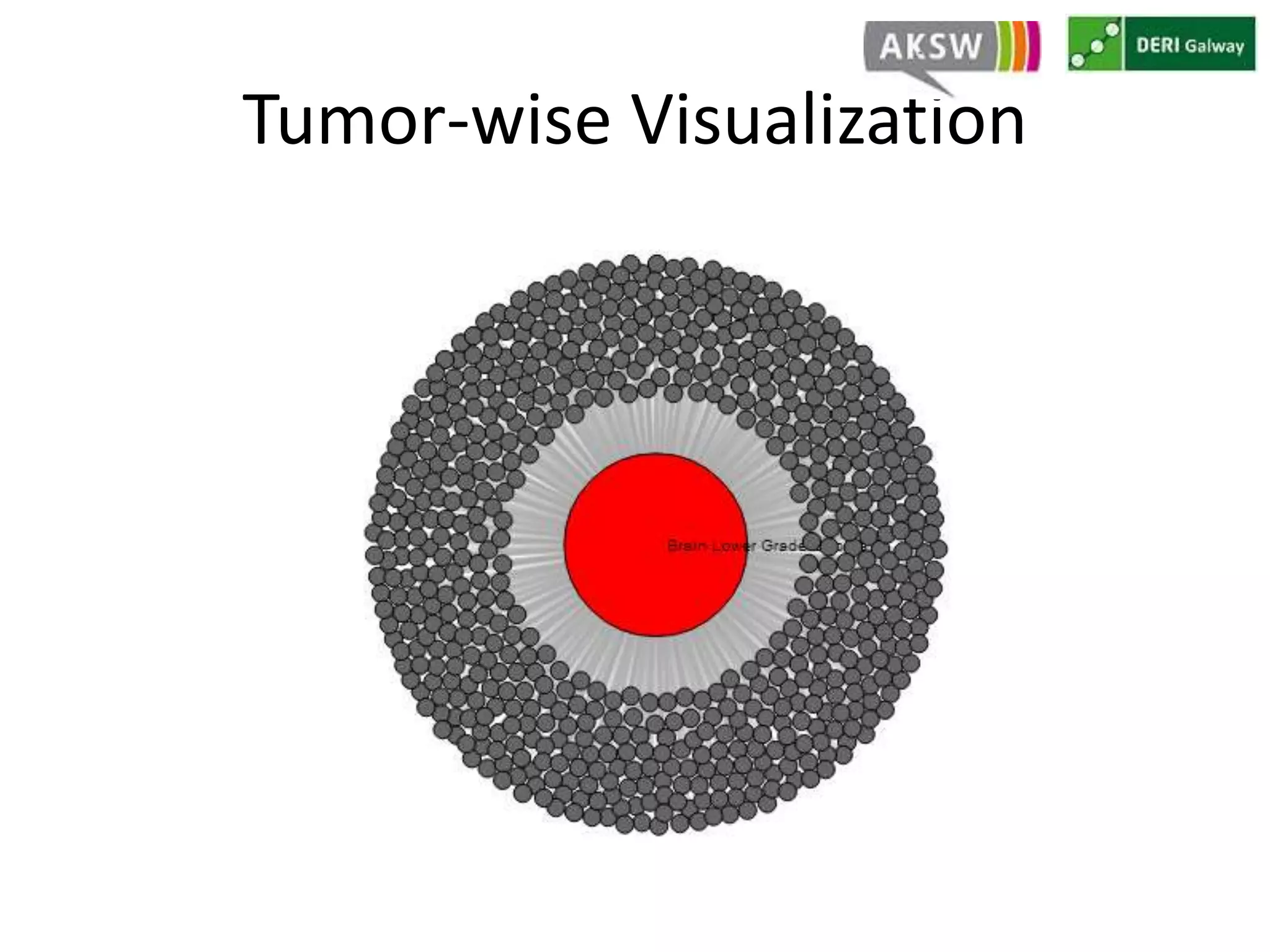
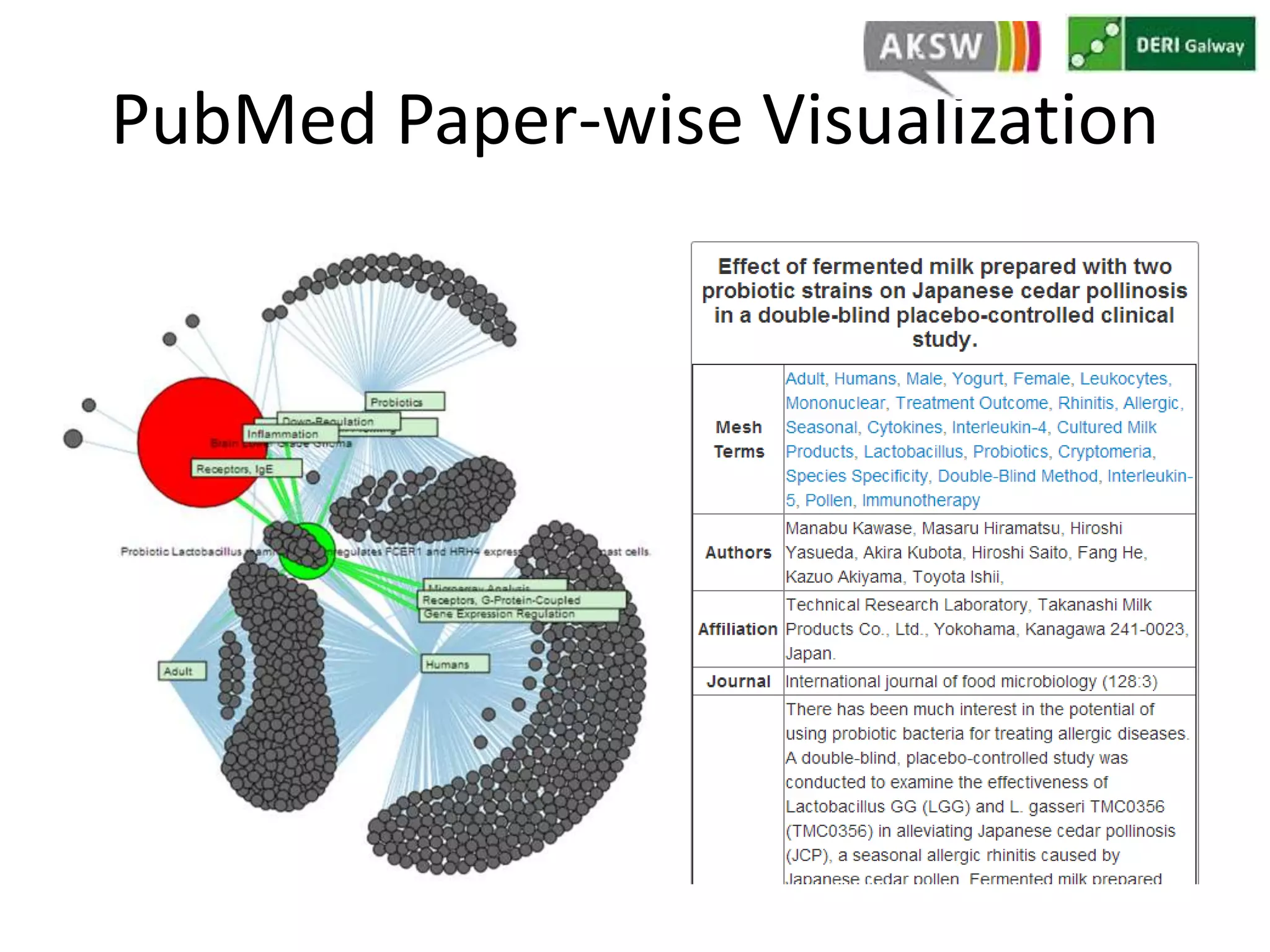
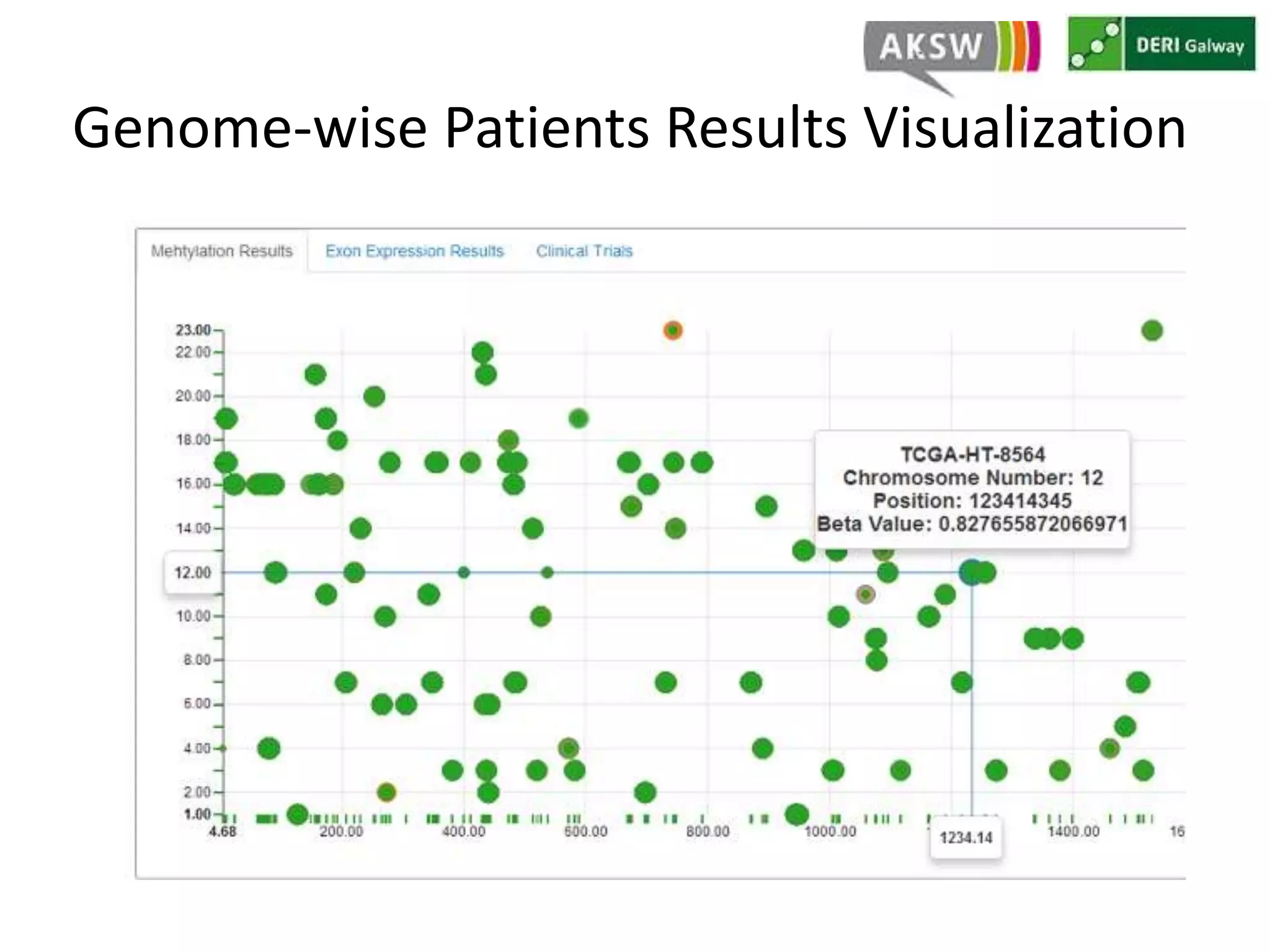
This document discusses fostering serendipity through linking large biomedical datasets. It linked over 30 billion triples from The Cancer Genome Atlas (TCGA) and over 23 million publications from PubMed. It developed an architecture called TopFed to continuously integrate new data through parallel querying. TopFed was evaluated against the FedX system and shown to have significantly better performance, with query runtimes over 75 times faster for some queries. A visualization interface was also created to explore the linked data.