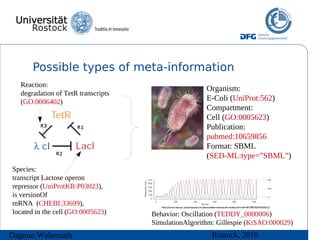
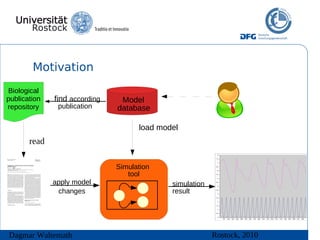
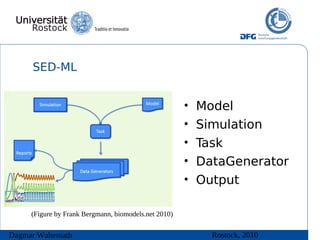
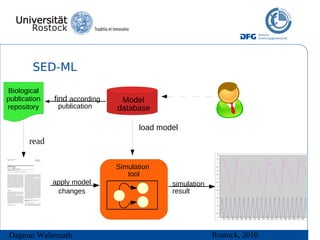
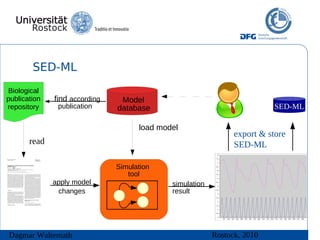
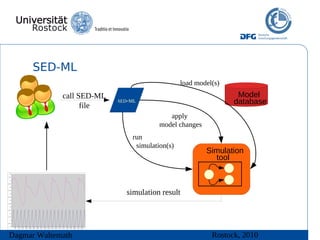
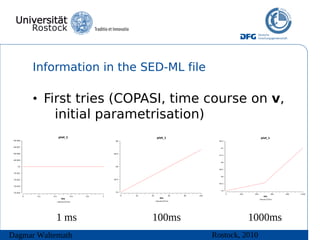
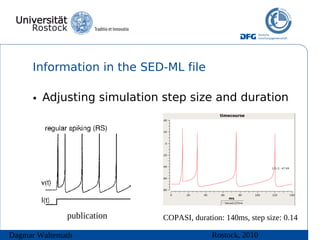
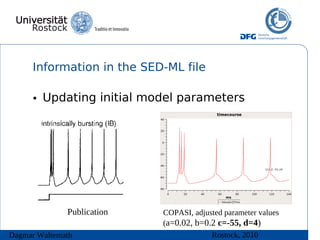
The document discusses model meta-information and simulation experiment descriptions. It describes how meta-information helps understand models by providing context through annotations using controlled vocabularies. Standards like MIRIAM ensure meta-information is computer-readable. Simulation Experiment Description Markup Language (SED-ML) allows storing and exchanging simulation experiments to facilitate model reuse. SED-ML specifications are under development and implementations exist in libraries and tools.

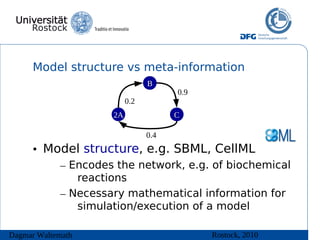
![Model structure vs meta-information
B
[SBO] 0.9 [Uniprot]
Protein complex 0.2 Tetracycline
[UniProt] repressor
[GO] 2A C Lactose protein
translation Operon
process 0.4 Repressor
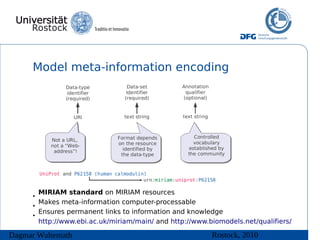
• Model meta-information helps “understanding” the
model
– MIRIAM (Minimum Information Requested in the
Annotation of Models)
– Use of controlled annotation, particularly ontologies,
including Gene Ontology, Systems Biology Ontology,
UniProt, CheBi ...
Dagmar Waltemath Rostock, 2010](https://image.slidesharecdn.com/waltemath-ontoworkshop-100527022429-phpapp02/85/Meta-Information-for-Bio-Models-5-320.jpg)