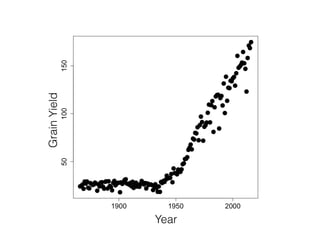
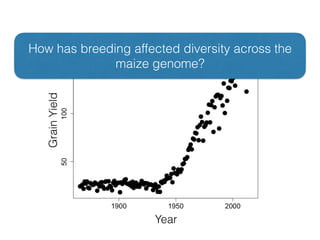
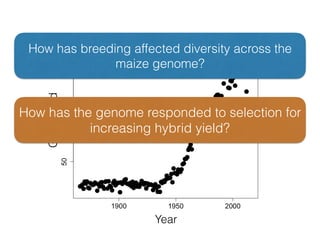
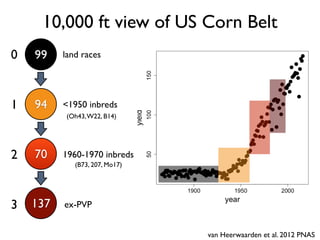
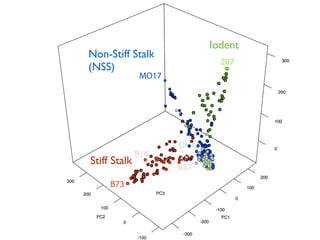
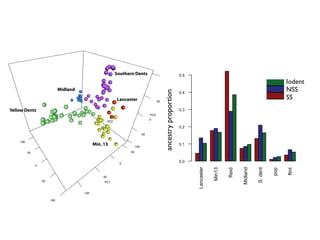
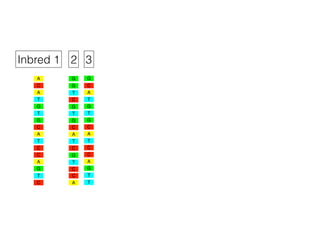
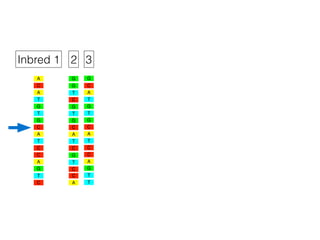
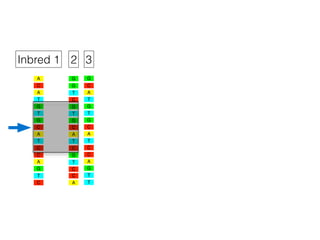
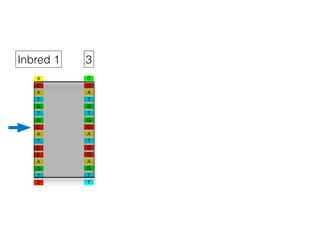
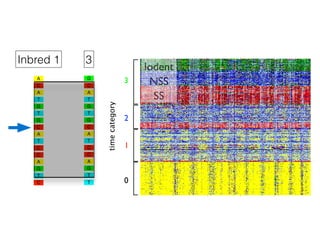
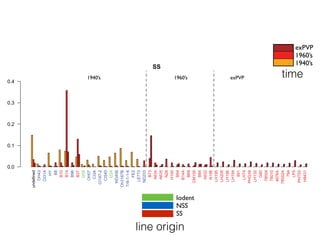
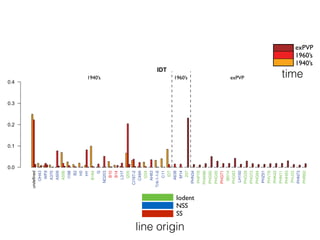
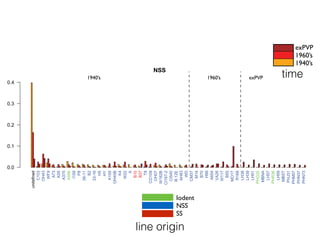
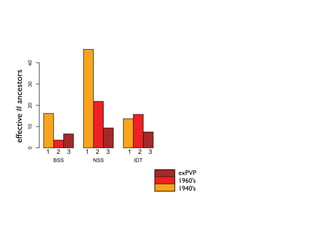
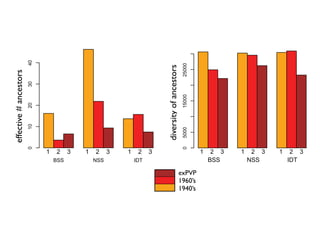
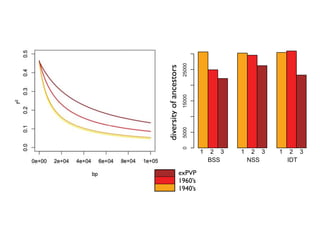
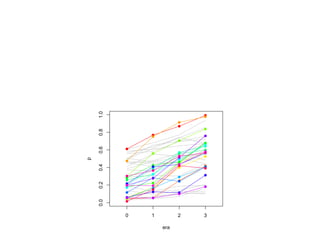
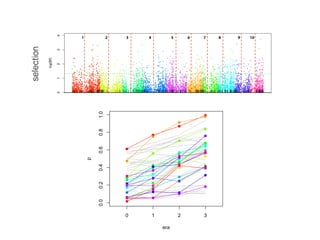
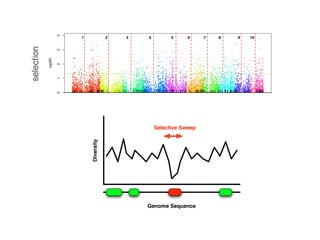
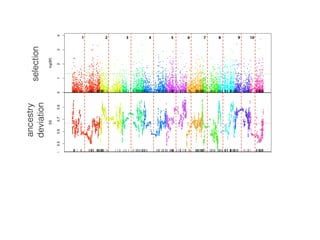
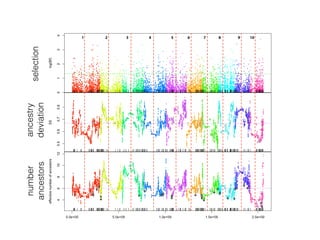
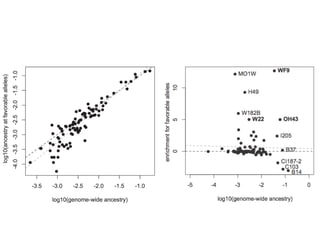
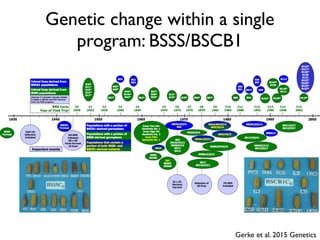
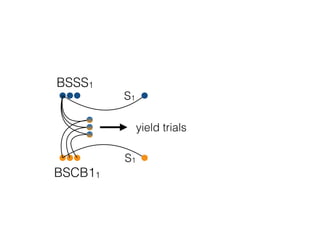
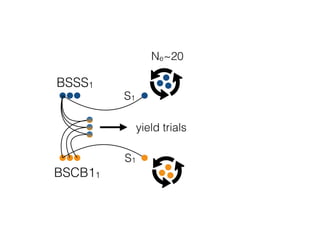
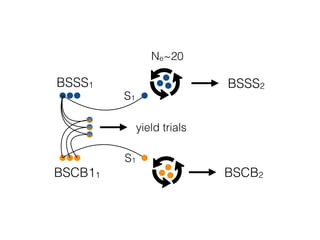
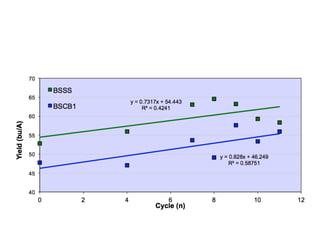
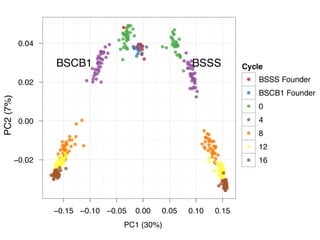
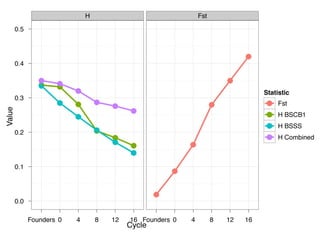
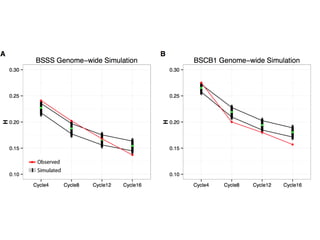
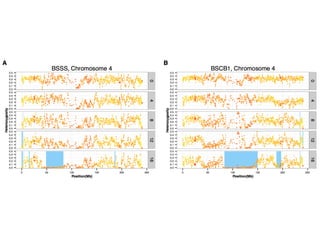
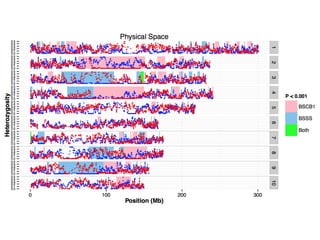
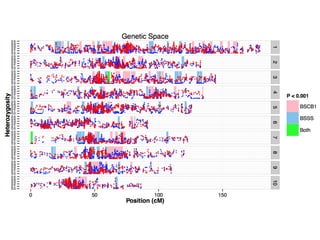
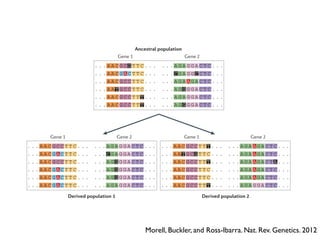
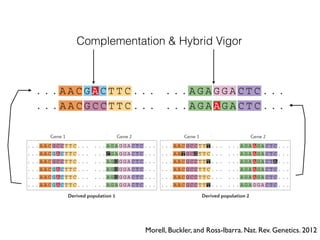
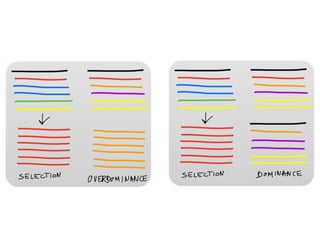
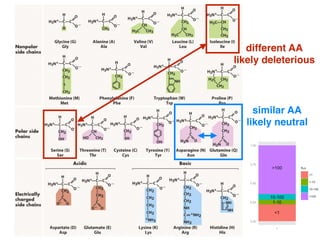
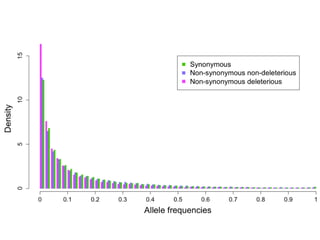
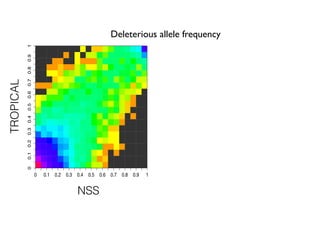
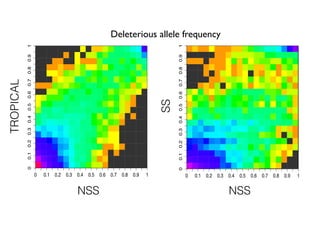
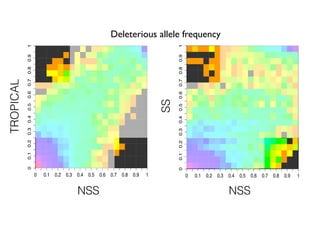
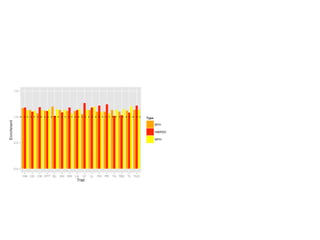
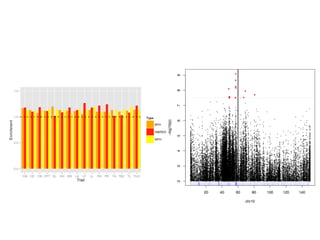
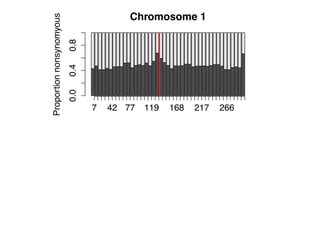
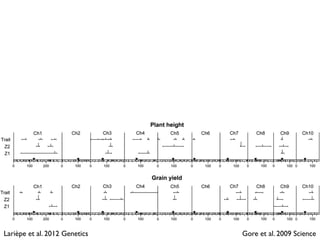
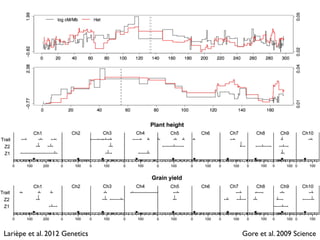
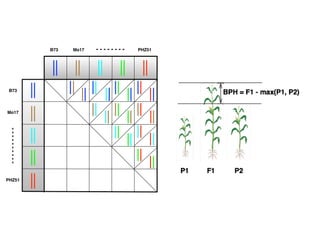
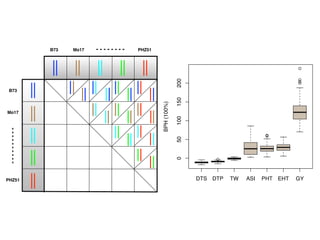
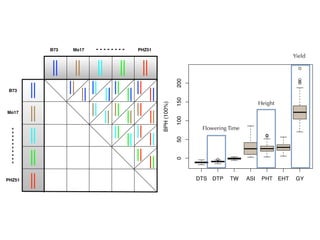
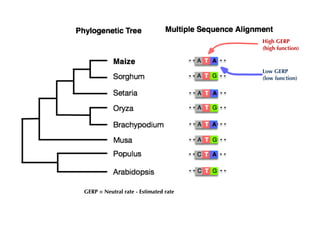
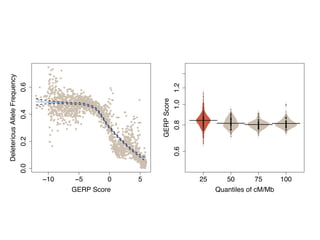
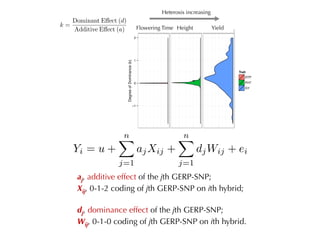
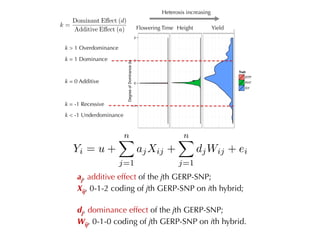
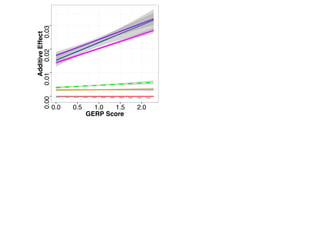
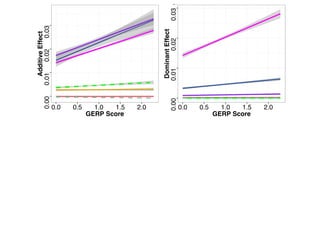
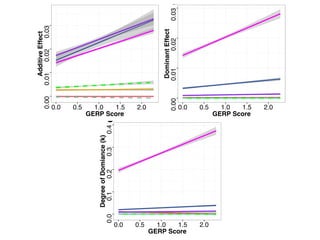
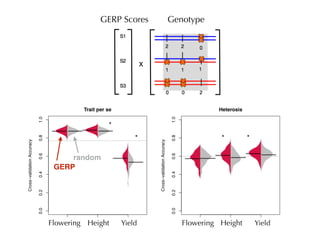
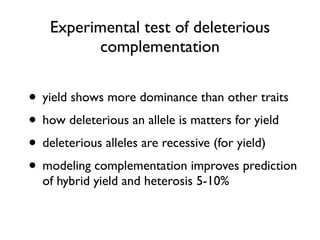
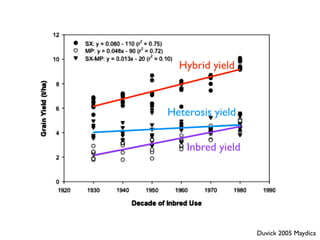
The document discusses how breeding has impacted genetic diversity in maize genomes over time. It examines changes in ancestry across the maize genome and how the genome has responded to selection for increasing hybrid yield. Specifically, it finds that changing ancestry, not selection sweeps, has driven diversity loss across heterotic groups. The diversity of ancestral lines making up modern inbreds has decreased as germplasm pools have become smaller and more homogeneous. Within a single breeding program, genetic drift in small breeding populations also reduced diversity despite selection for yield.