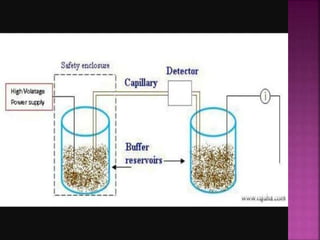
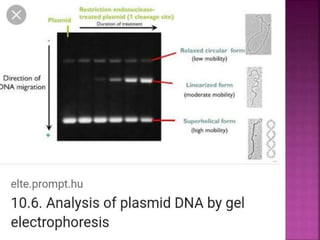
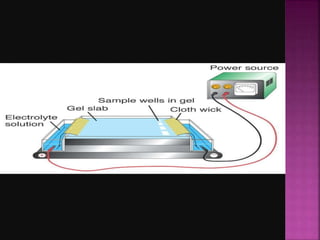
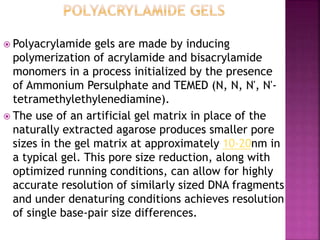
Electrophoresis is a laboratory technique used to separate macromolecules such as DNA and RNA based on size, with variations including gel electrophoresis and capillary electrophoresis. The method capitalizes on the charge of the molecules in the presence of an electric field, allowing for practical applications in genetic analysis, forensic science, and ink analysis. Key methods like agarose and polyacrylamide gel electrophoresis offer different resolutions and are essential in various biochemical analyses.