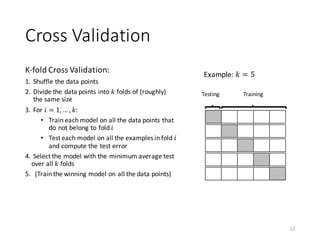
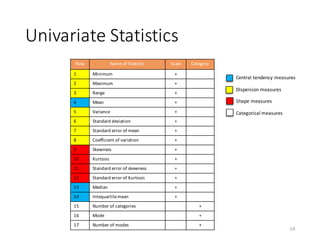
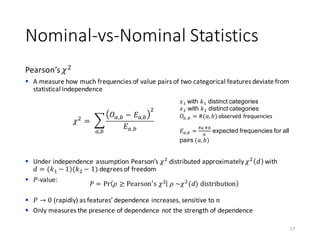
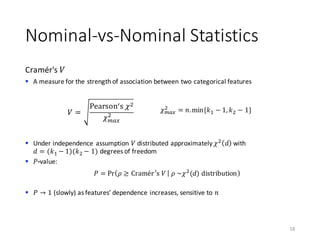
The document outlines the processes involved in data preparation, including pre-processing and descriptive statistics using SystemML. It covers various aspects such as the transformation of input data, training and testing data sets, and detailed statistical measures for analyzing data relationships. The procedures and built-in functions for handling categorical variables, cross-validation methods, and criteria for descriptive statistics are also discussed.




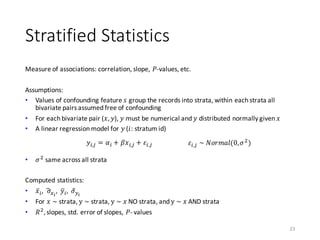
![Transform Specification
§ Transformations operate on individual columns
§ All required transformations specified in a JSON file
§ Property na.strings in the mtd file specifies missing values
Example:
data.spec.json data.csv.mtd
6
{
"data_type": "frame",
"format": "csv",
"sep": ",",
"header": true,
"na.strings": [ "NA", "" ]
}
{
“ids": true
, "omit": [ 1, 4, 5, 6, 7, 8, 9 ]
, "impute":
[ { “id": 2, "method": "constant",
"value": "south" }
,{ “id": 3, "method":
"global_mean" }
]
,"recode": [ 1, 2, 4, 5, 6, 7 ]
,"bin":
[ { “id": 8, "method": "equi-
width", "numbins": 3 } ]
,"dummycode": [ 2, 5, 6, 7, 8, 3 ]
}](https://image.slidesharecdn.com/s2datapreparationanddescriptivestatisticsinsystemml-160913182813/85/Data-preparation-training-and-validation-using-SystemML-by-Faraz-Makari-Manshadi-6-320.jpg)





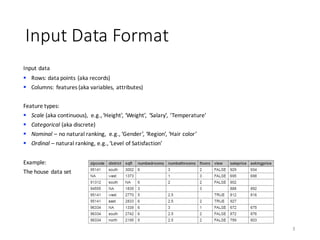
![Scale-vs-Scale Statistics
Pearson’s correlation coefficient
§ A measure of linear dependence between scale features
§ 𝜌)
measures accuracy of 𝑥) ~ 𝑥0
16
𝜌 =
123(56,57)
9:69:7
, 𝜌 ∈ [−1,+1]
1 − 𝜌)
=
∑ 𝑥A,) − 𝑥BA,)
)C
AD0
∑ 𝑥A,) − 𝑥̅A,)
)C
AD0
Residual Sum of Squares (RSS)
Total Sum of Squares (TSS)](https://image.slidesharecdn.com/s2datapreparationanddescriptivestatisticsinsystemml-160913182813/85/Data-preparation-training-and-validation-using-SystemML-by-Faraz-Makari-Manshadi-16-320.jpg)
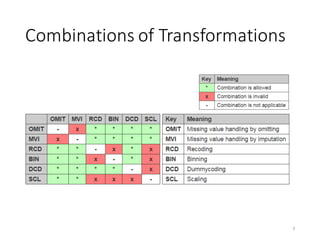
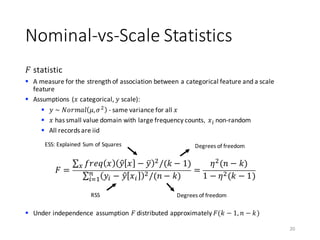
![Nominal-vs-Scale Statistics
Eta statistic
§ A measure for the strength of association between a categorical feature and a scale
feature
§ 𝜂)
measures accuracy of 𝑦 ~ 𝑥 similar to 𝑅)
statistic of linear regression
19
𝜂)
= 1 −
∑ 𝑦A − 𝑦B[𝑥A] )C
AD0
∑ 𝑦A − 𝑦k )C
AD0
RSS
TSS
𝑥 categorical
𝑦 scale
𝑦B[𝑥A]: average of 𝑦A among all records with
𝑥A = 𝑥](https://image.slidesharecdn.com/s2datapreparationanddescriptivestatisticsinsystemml-160913182813/85/Data-preparation-training-and-validation-using-SystemML-by-Faraz-Makari-Manshadi-19-320.jpg)
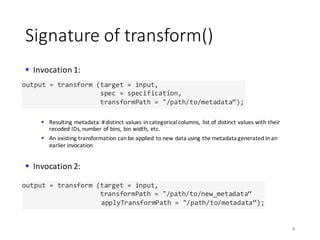
![Ordinal-vs-Ordinal Statistics
Spearman’s rank correlation coefficient
§ A measure for the strength of association between two ordinal features
§ Pearson’s correlation efficient applied to feature with values replaced by their ranks
Example:
21
8x
3)
11z
8{
5|
20
𝑥′
8
3
11
8
5
2
𝑥
4.5
2
6
4.5
3
1
𝑟
𝜌 =
123 (•6,•7)
9‚69‚7
𝜌 ∈ [−1, +1]](https://image.slidesharecdn.com/s2datapreparationanddescriptivestatisticsinsystemml-160913182813/85/Data-preparation-training-and-validation-using-SystemML-by-Faraz-Makari-Manshadi-21-320.jpg)