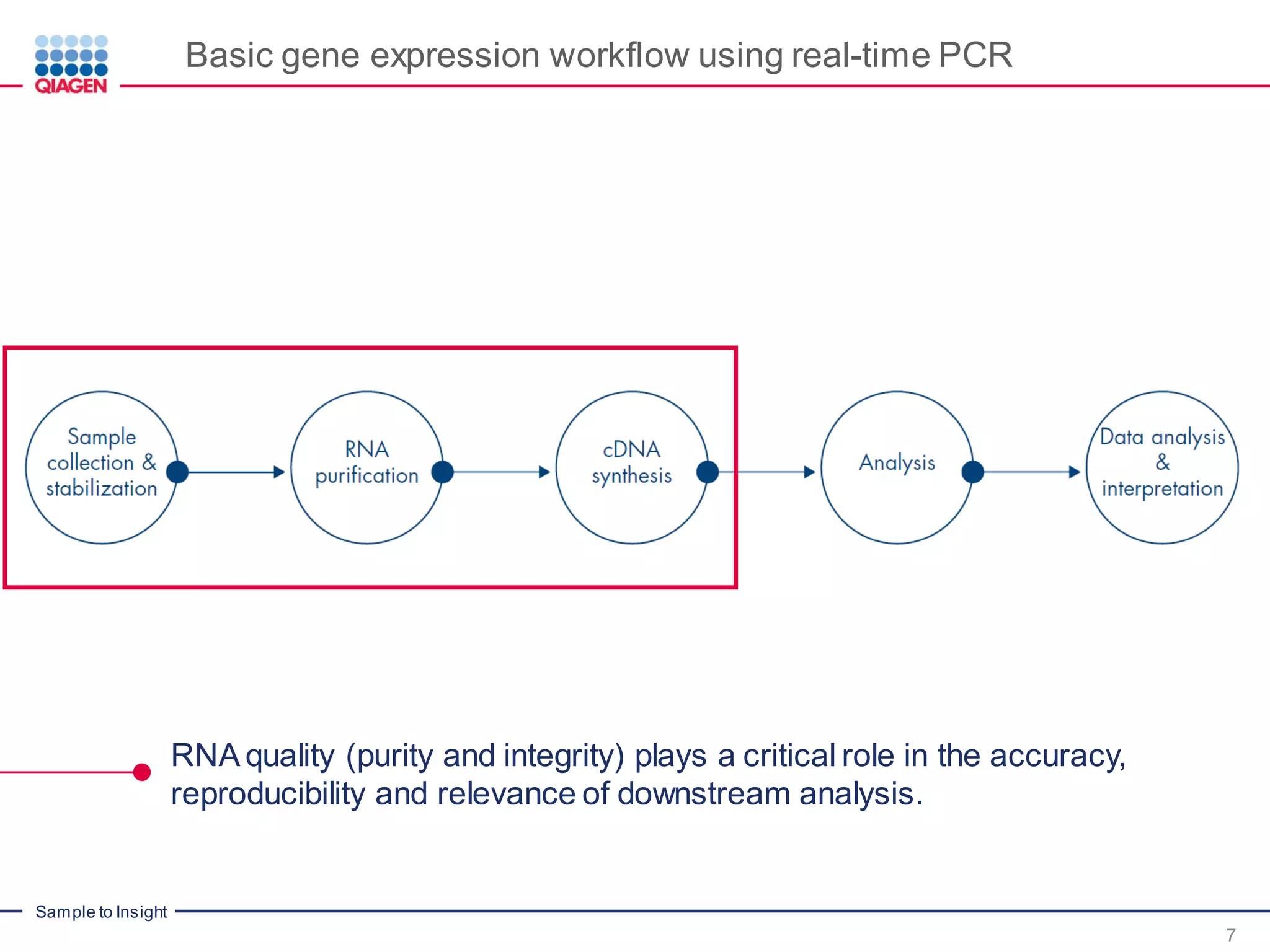
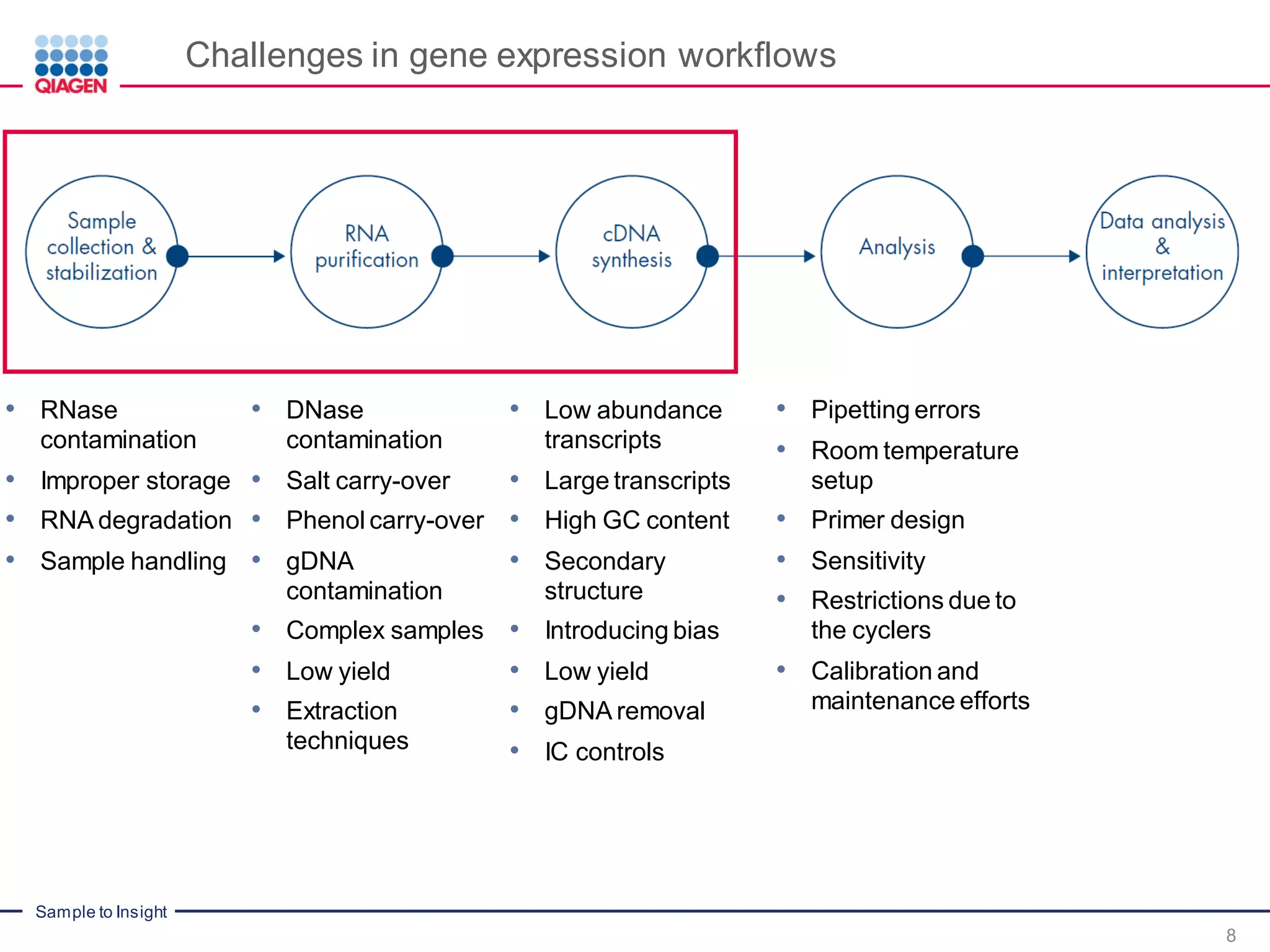
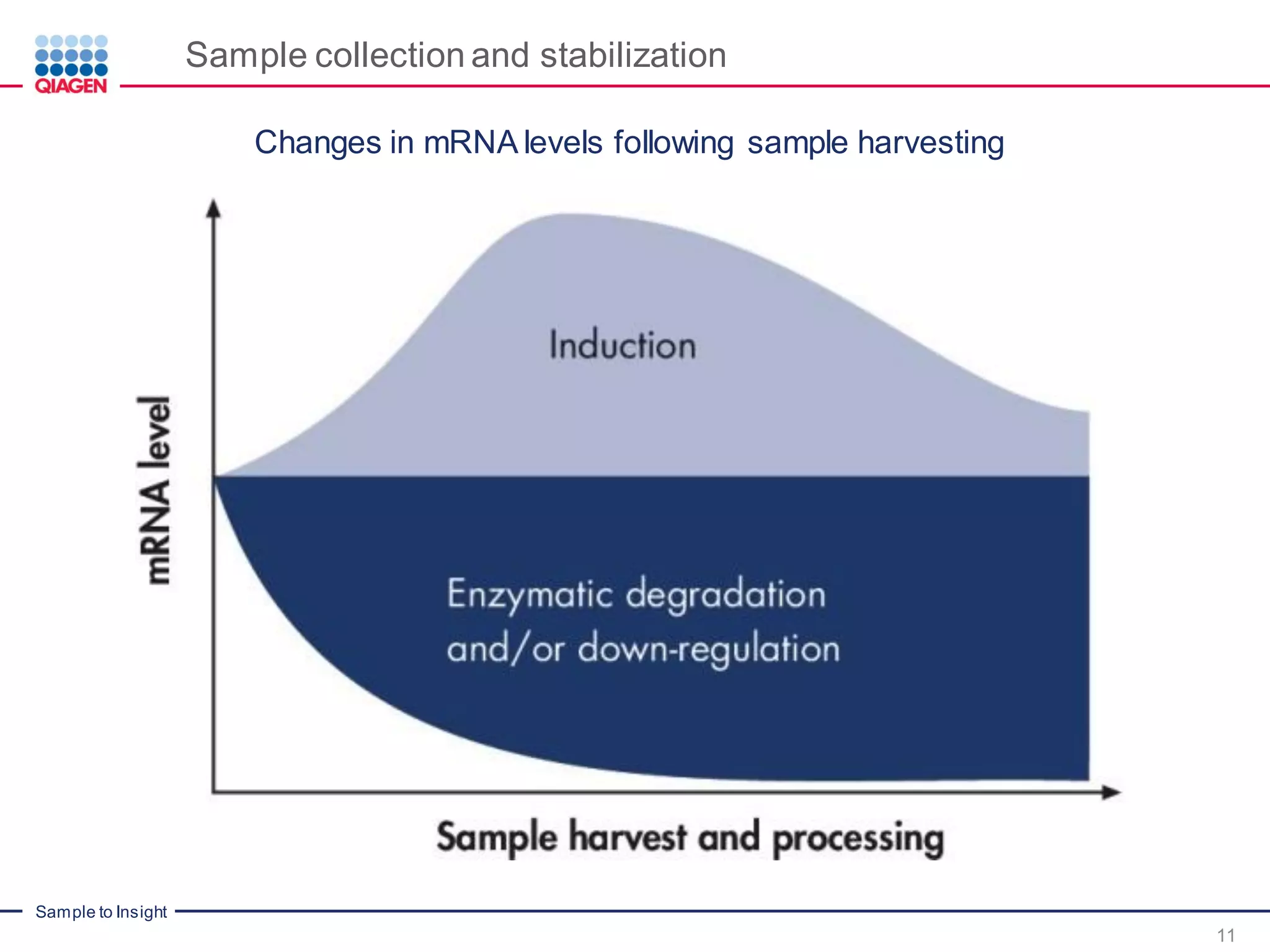
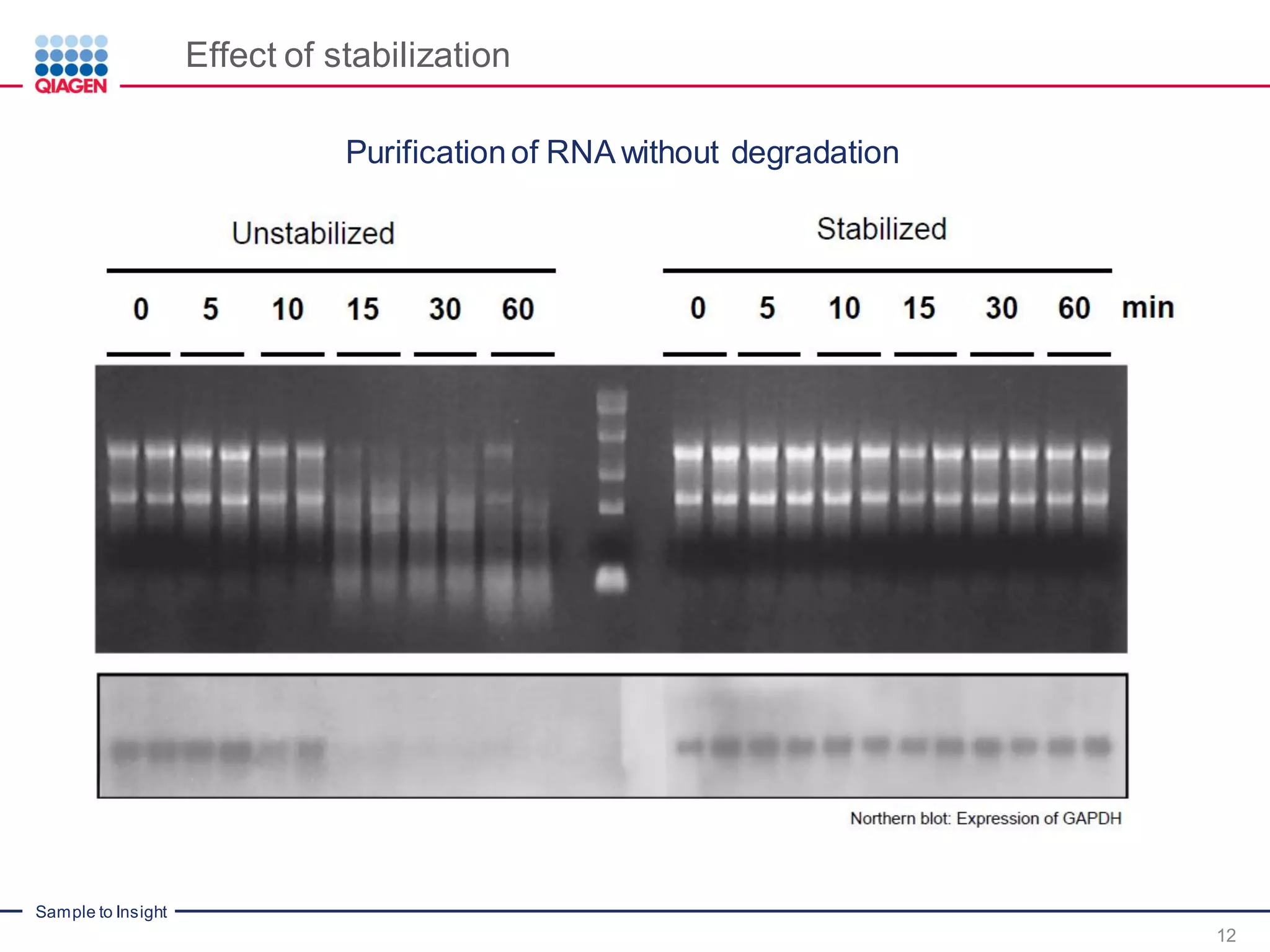
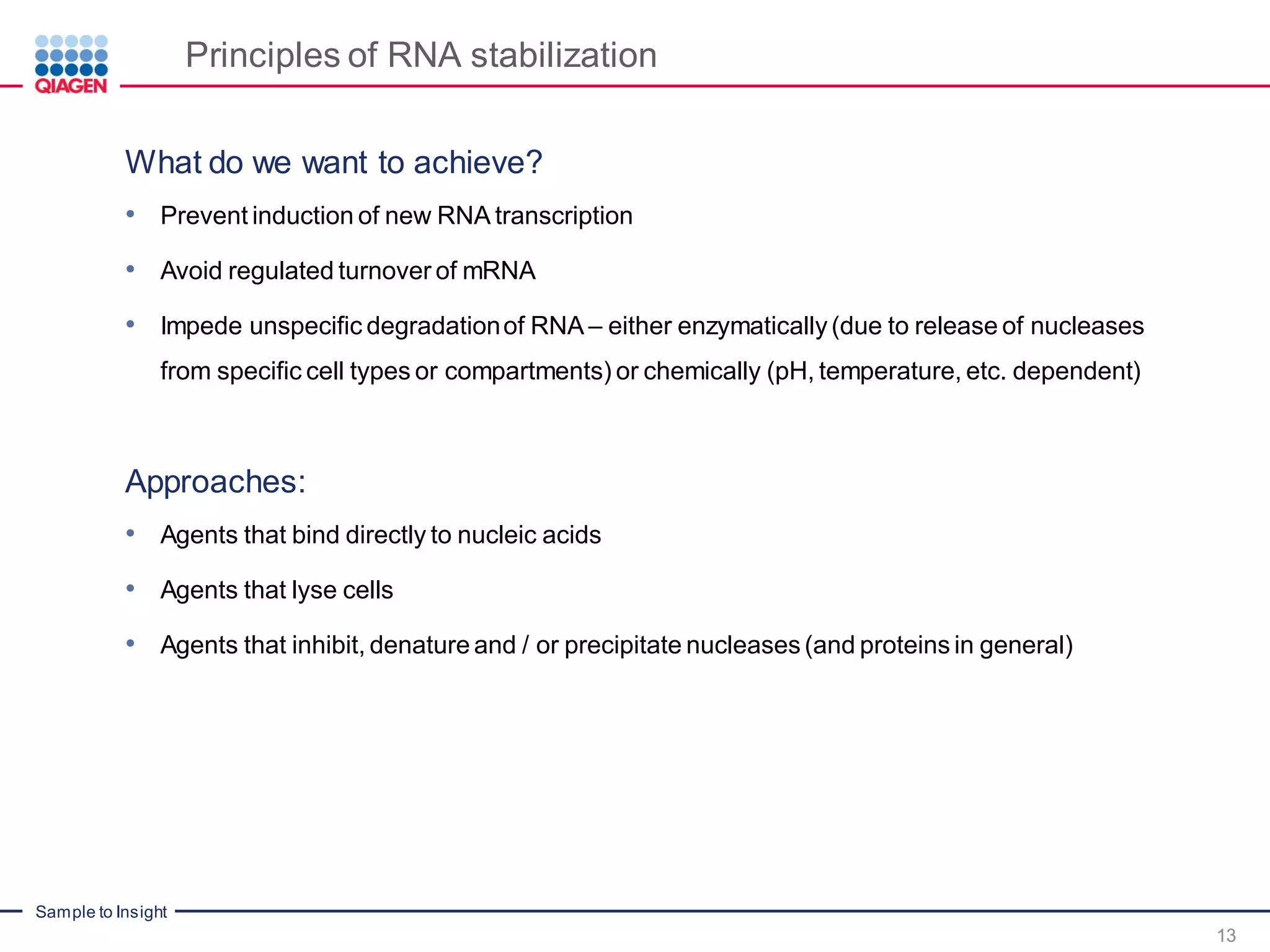
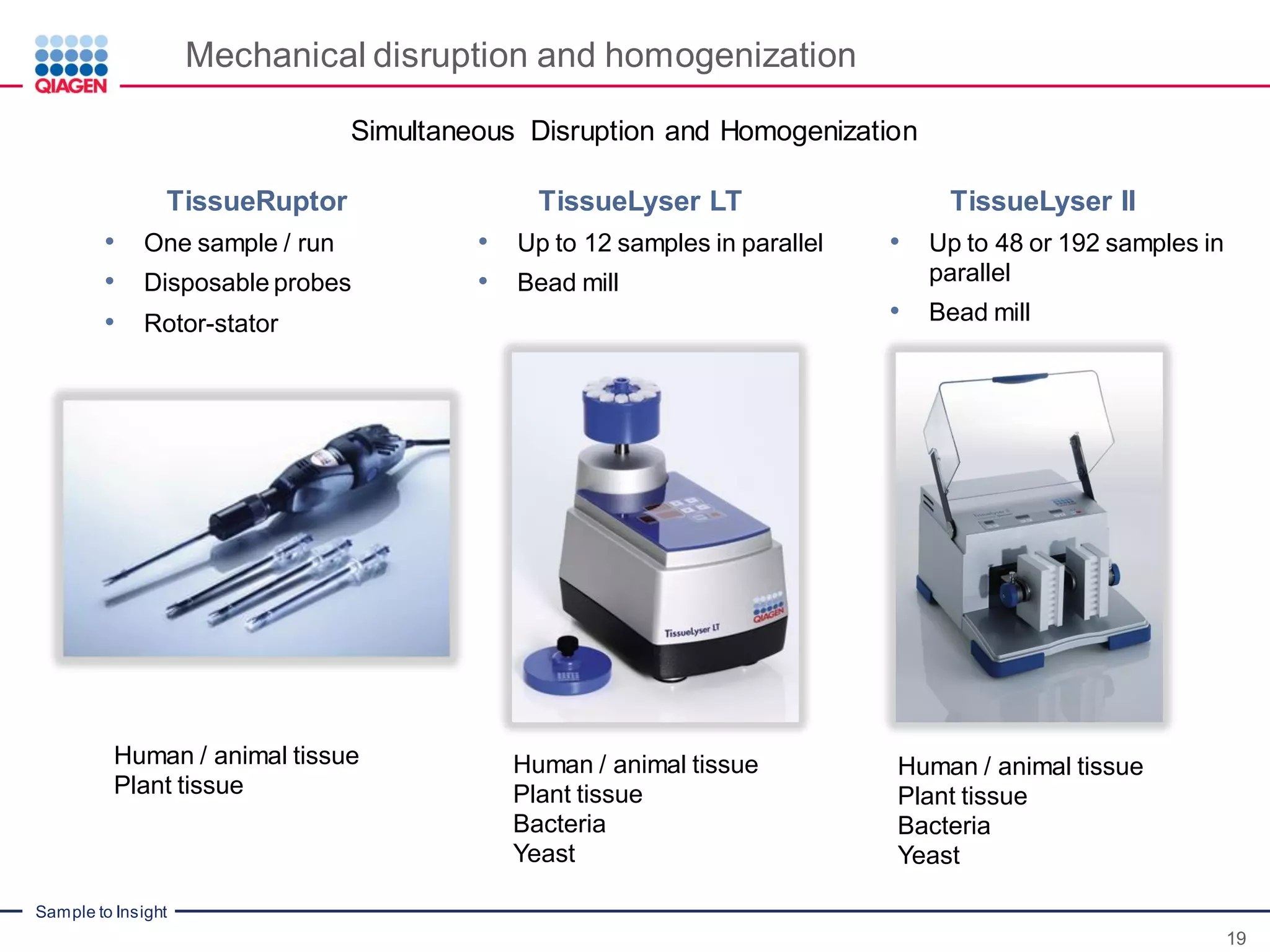
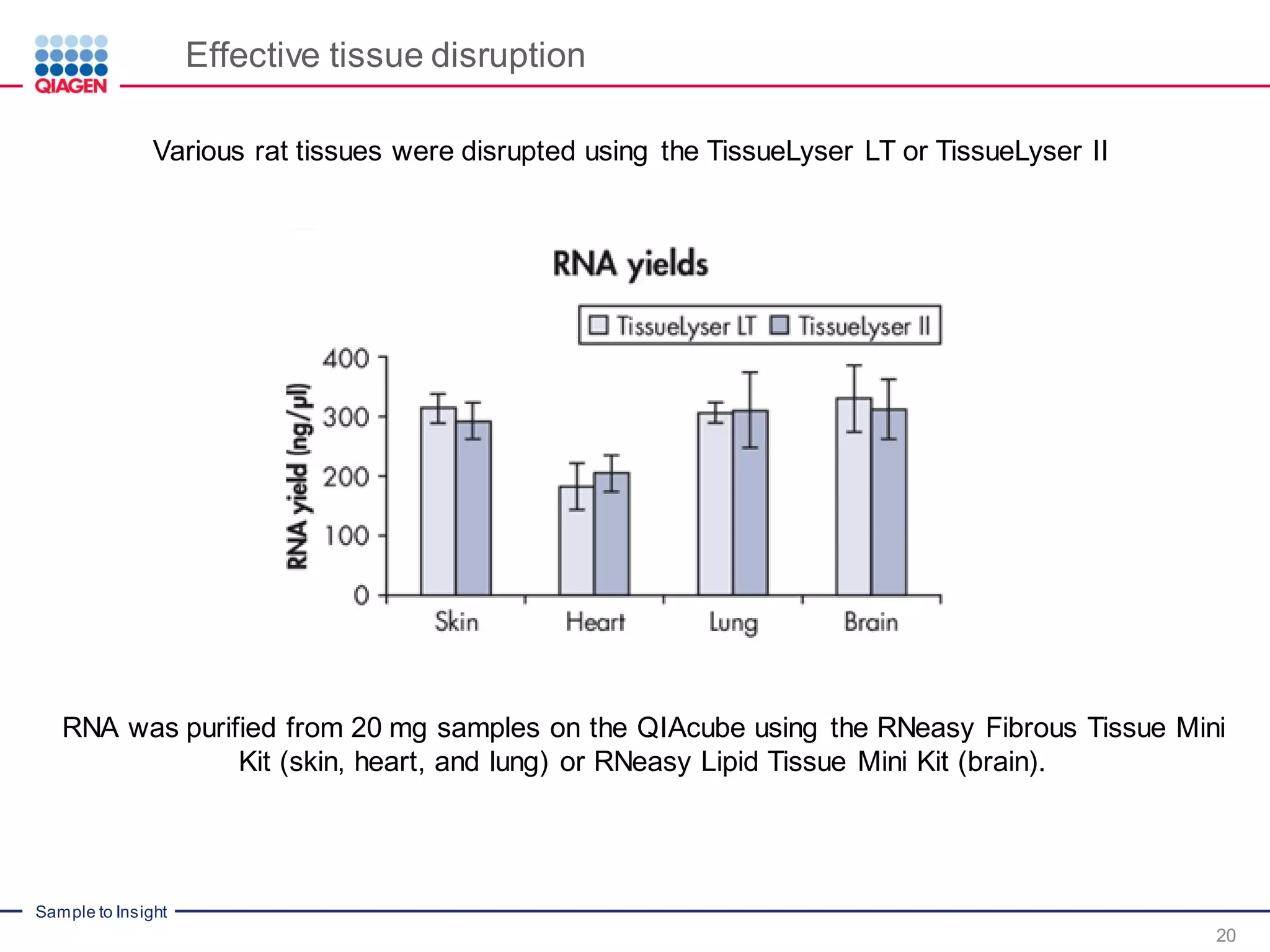
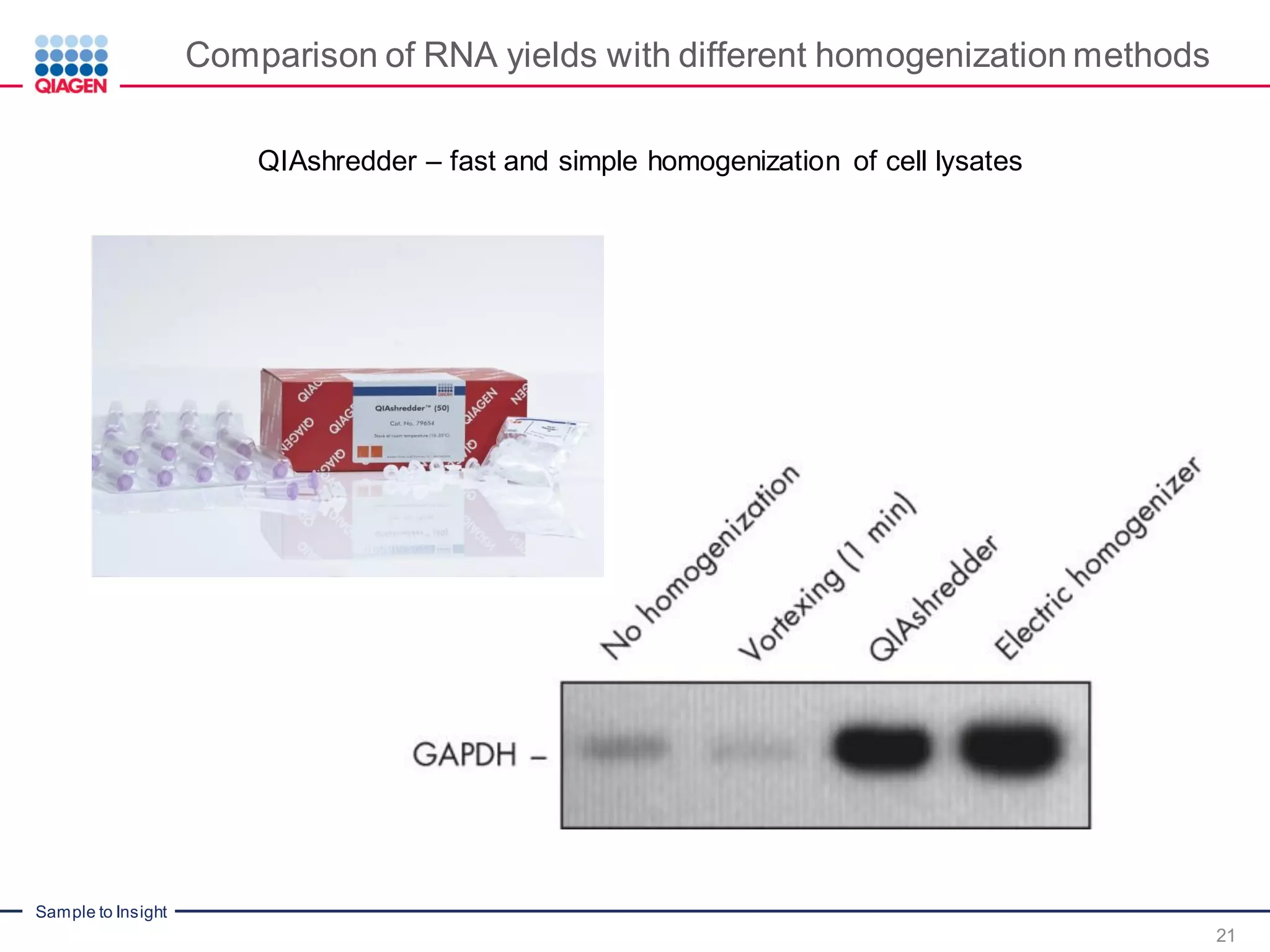
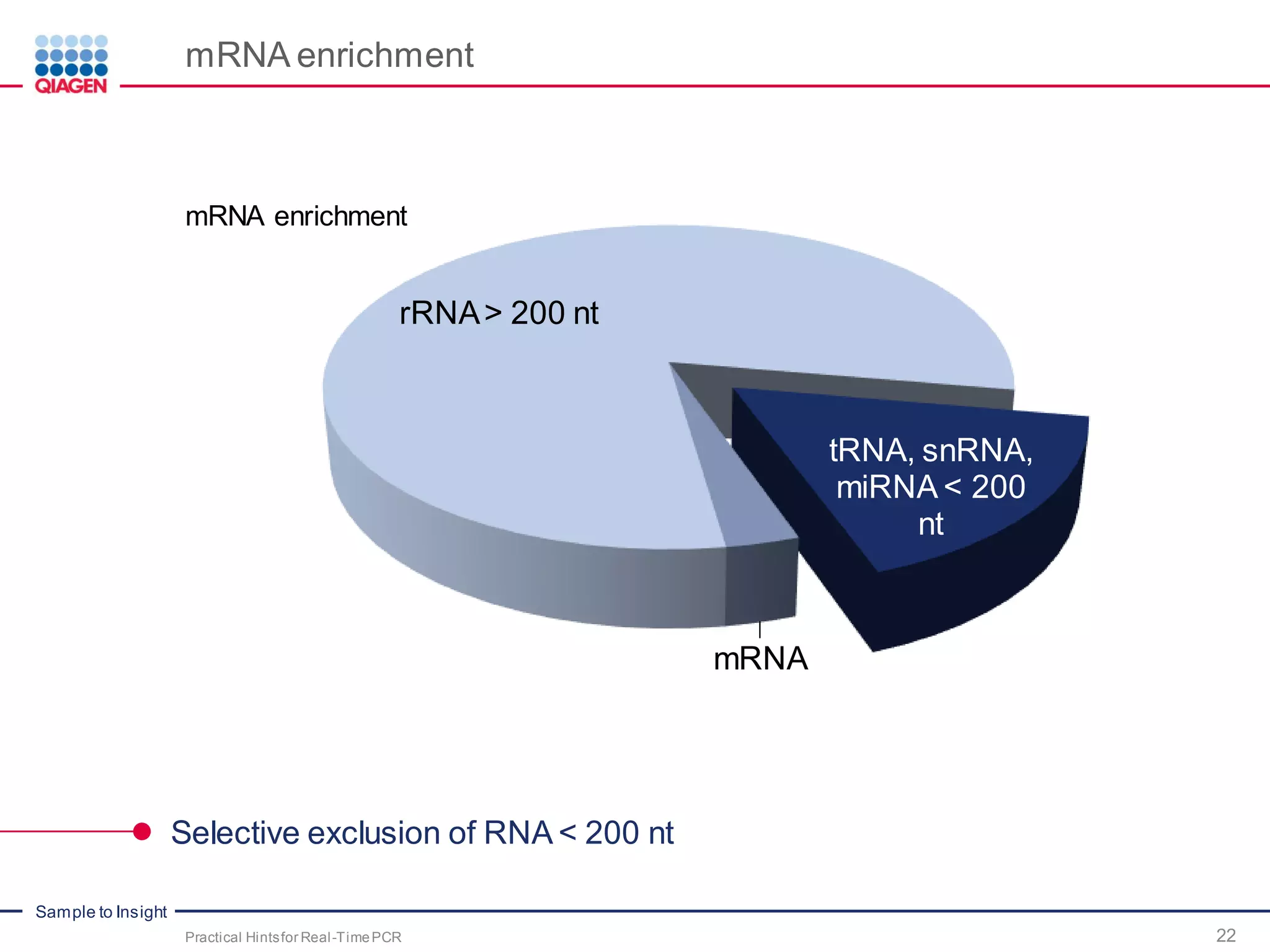
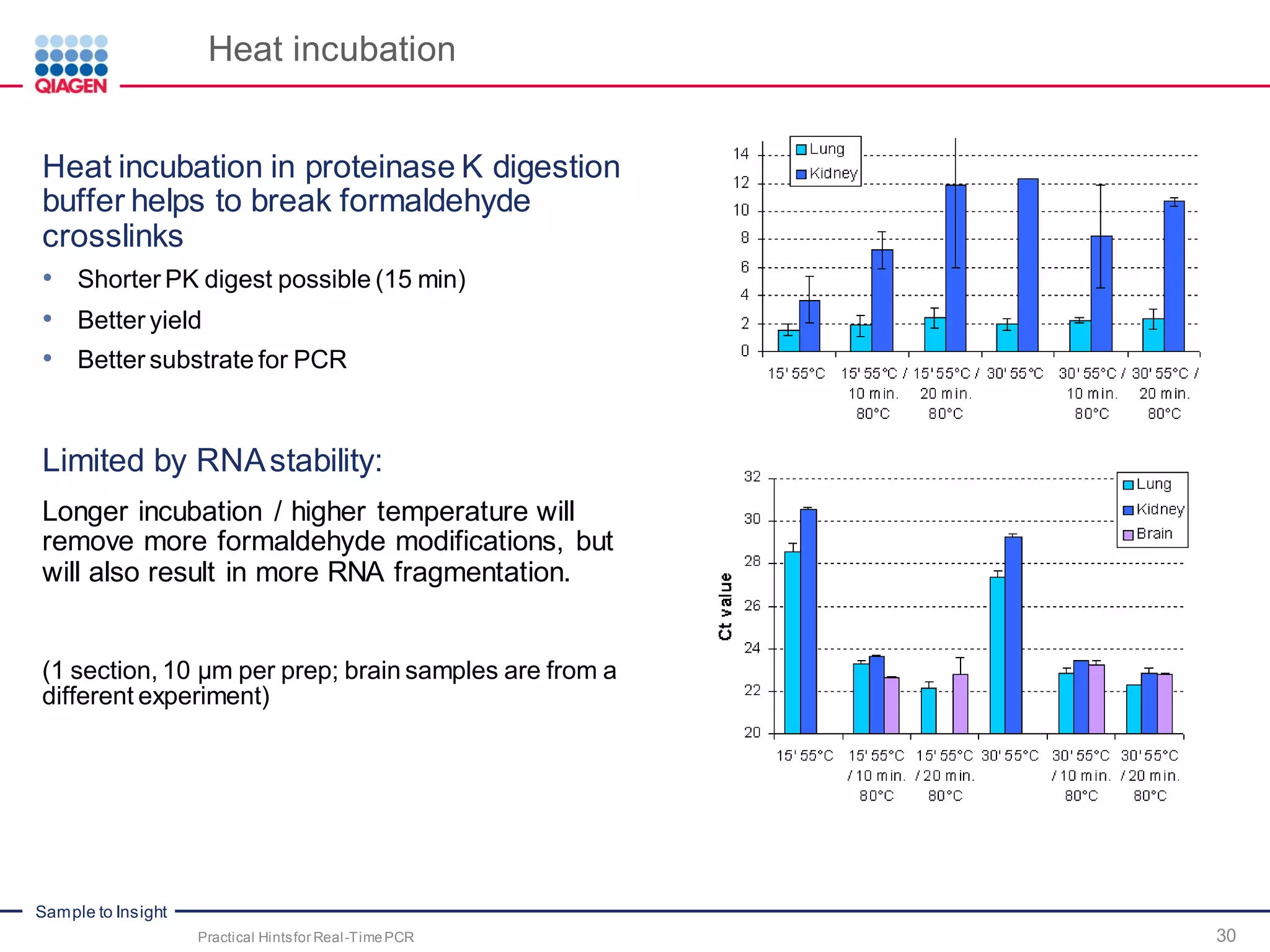
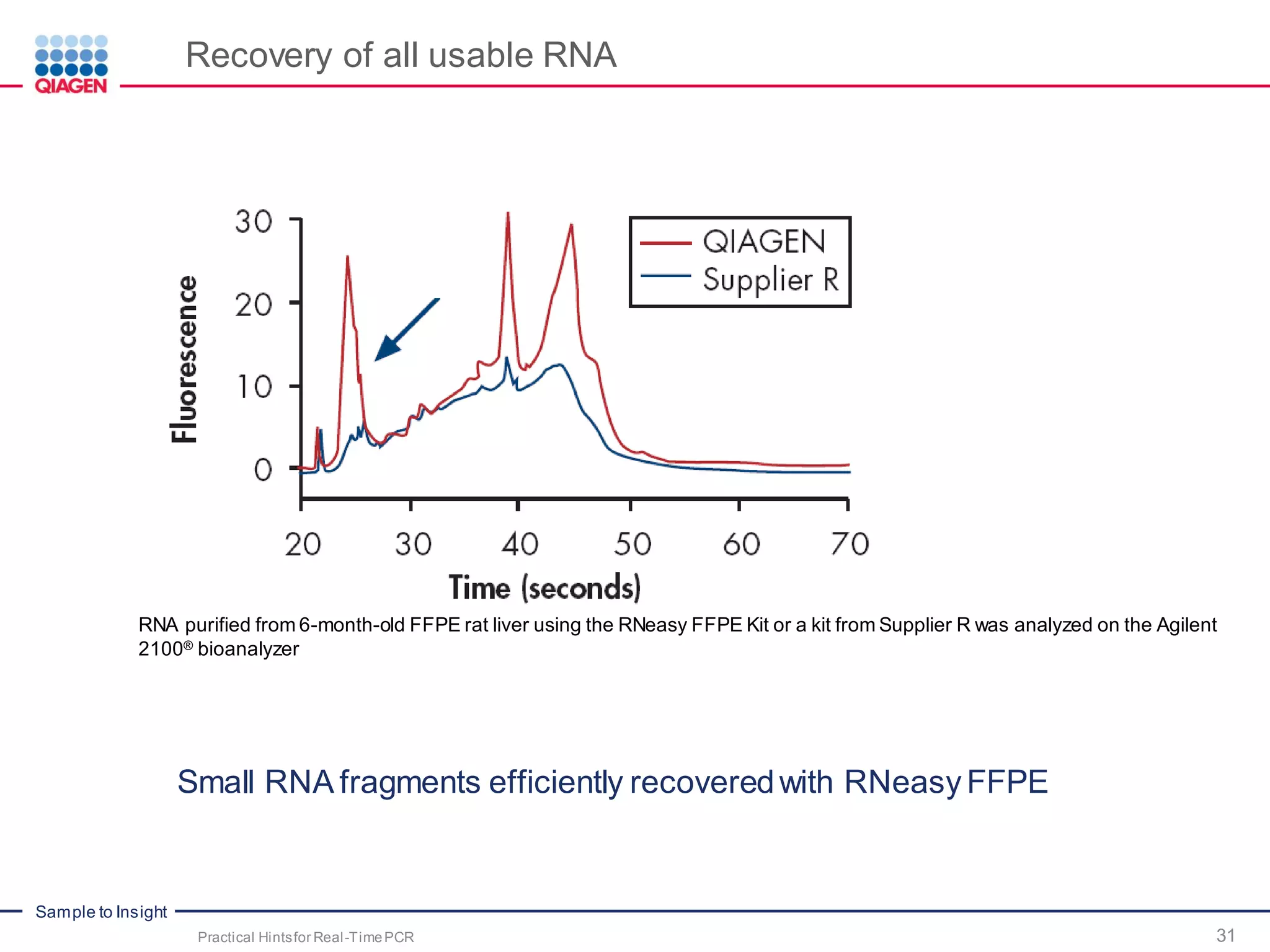
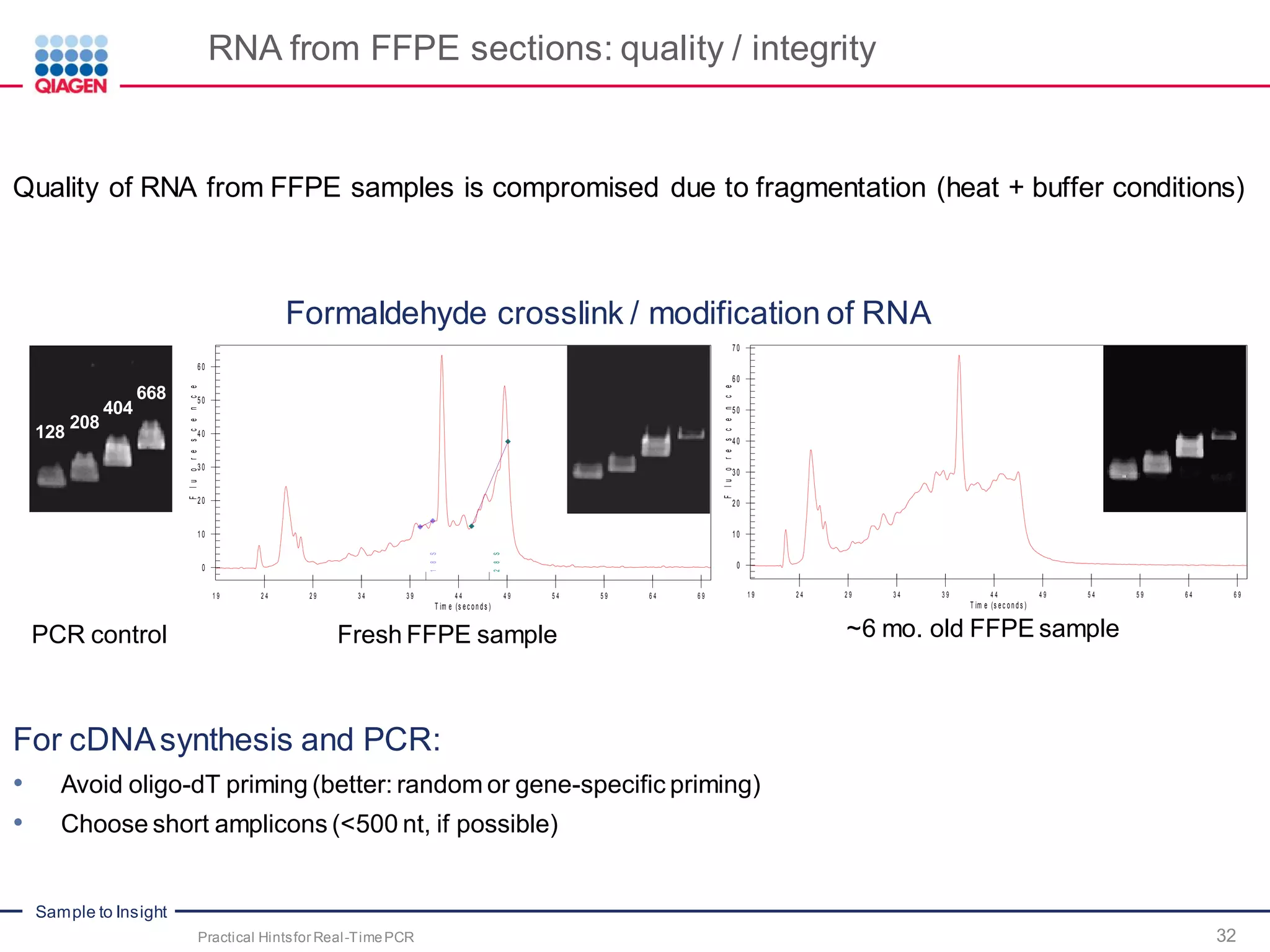
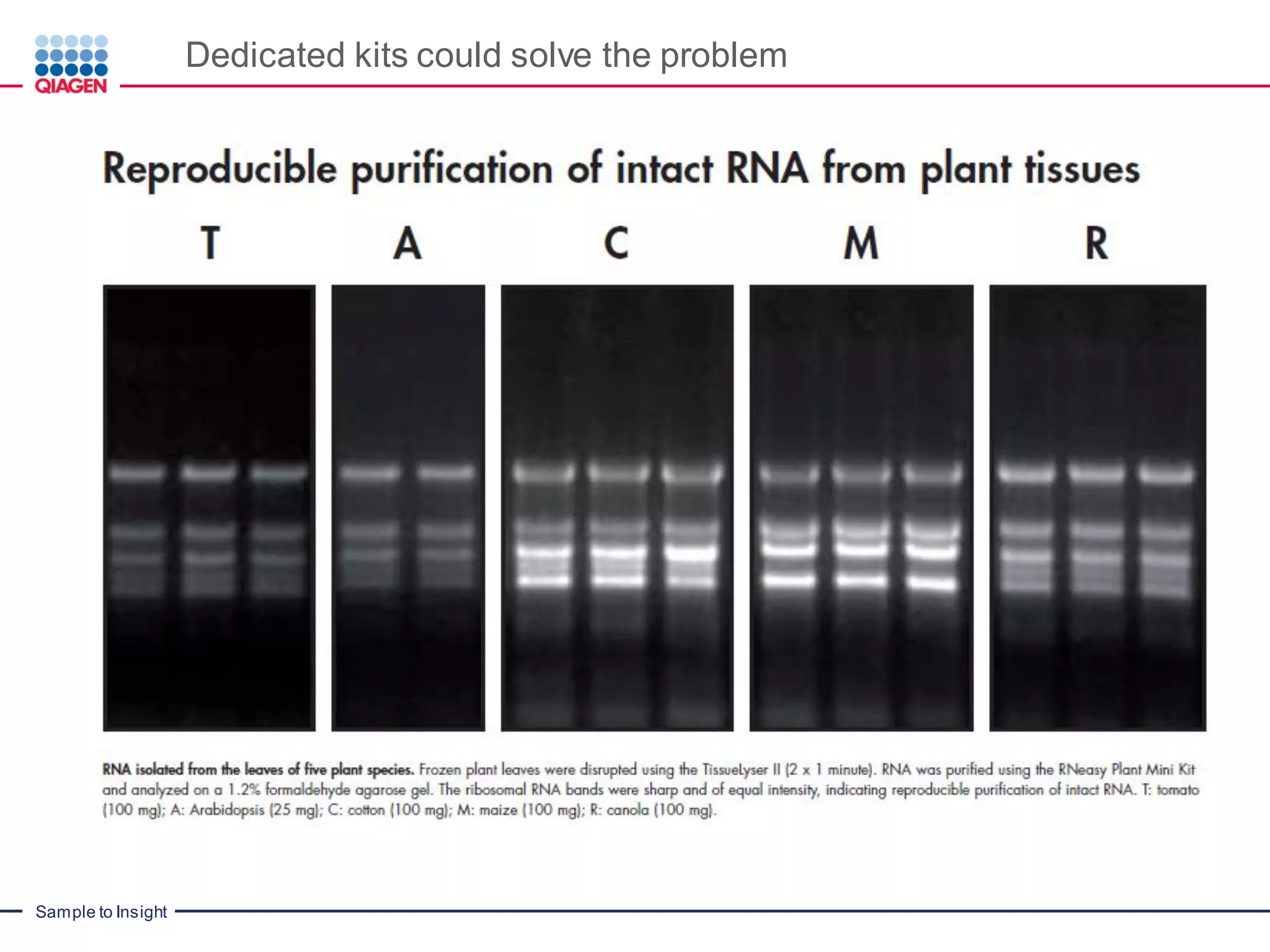
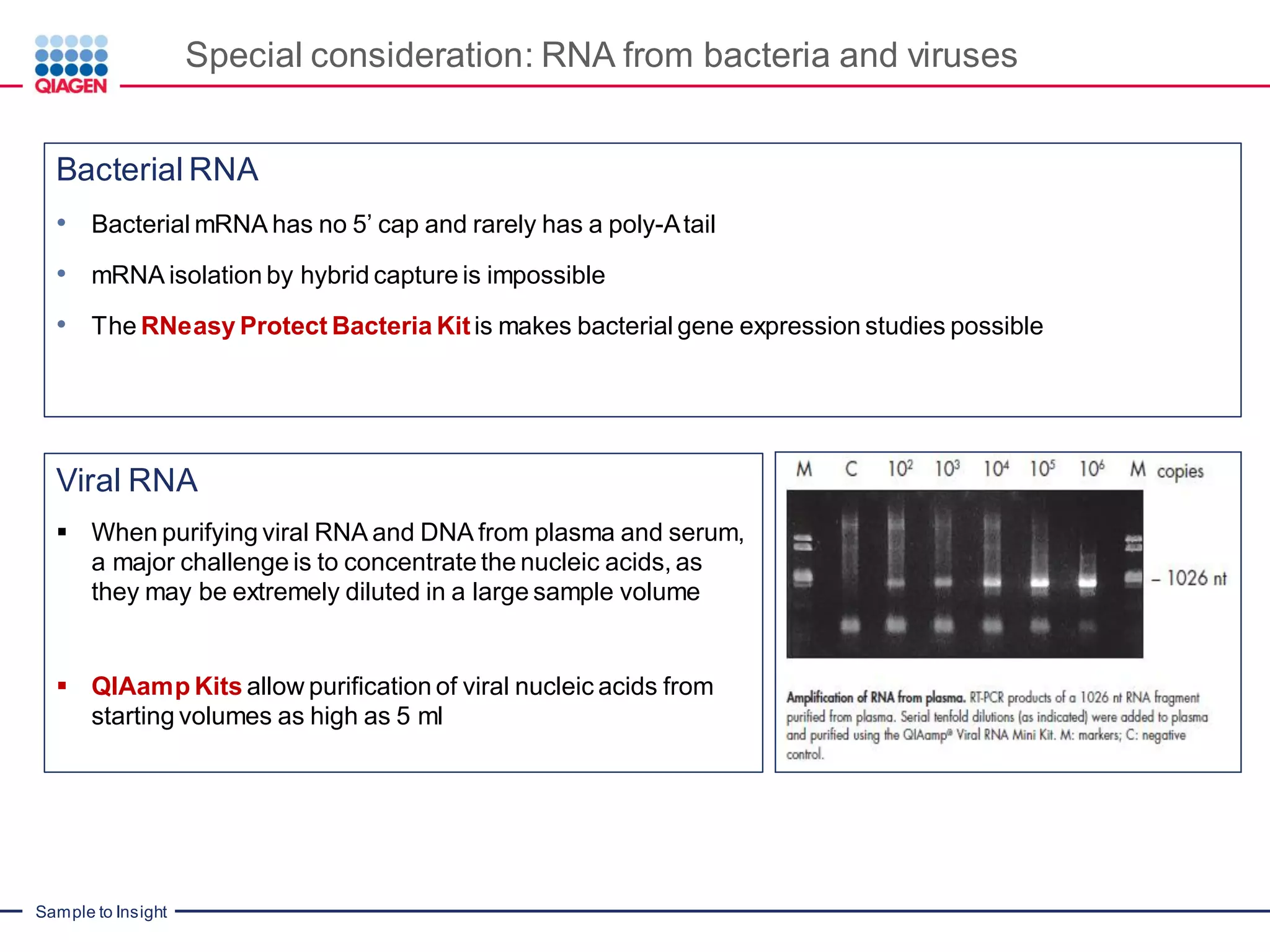
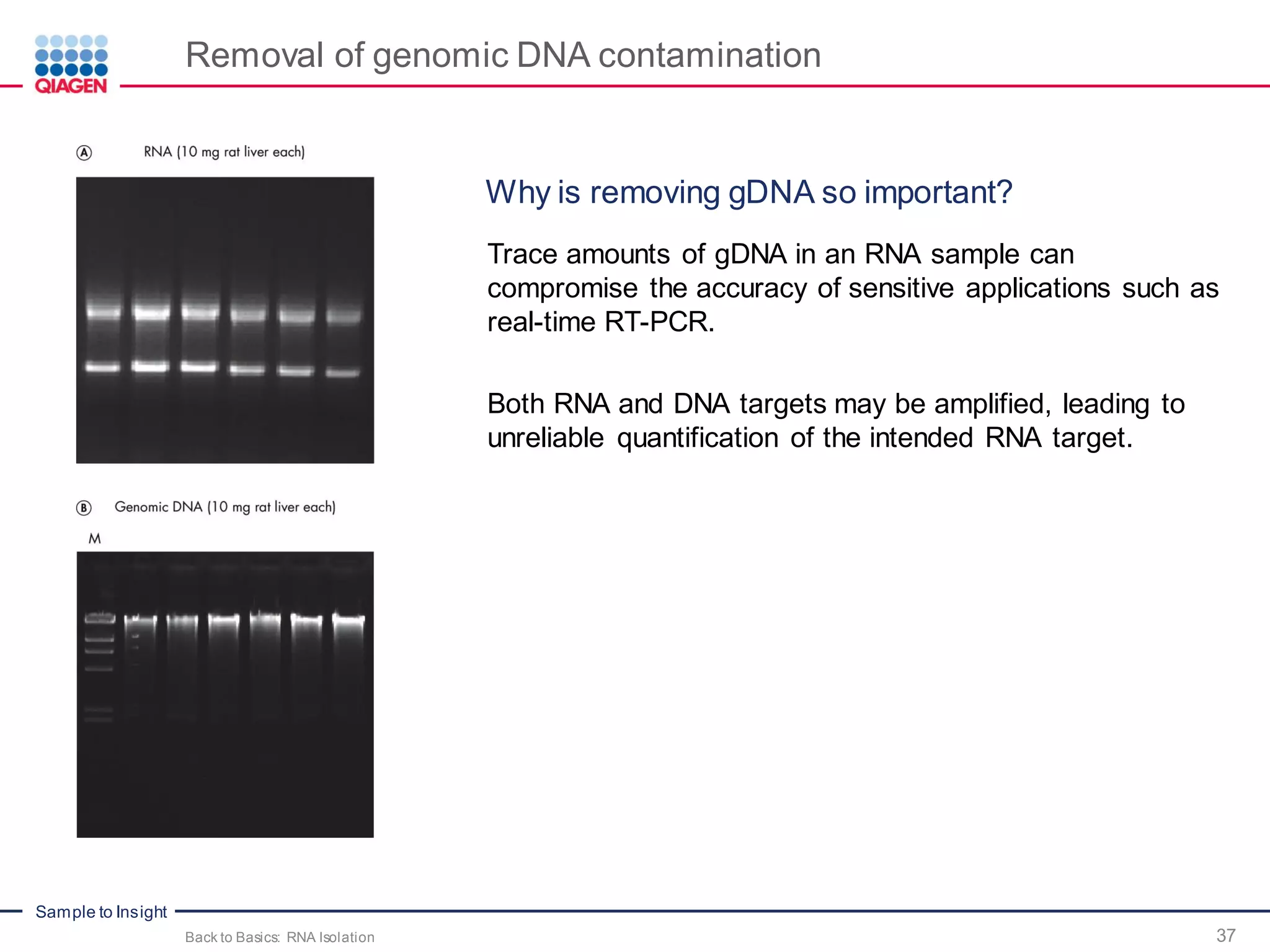
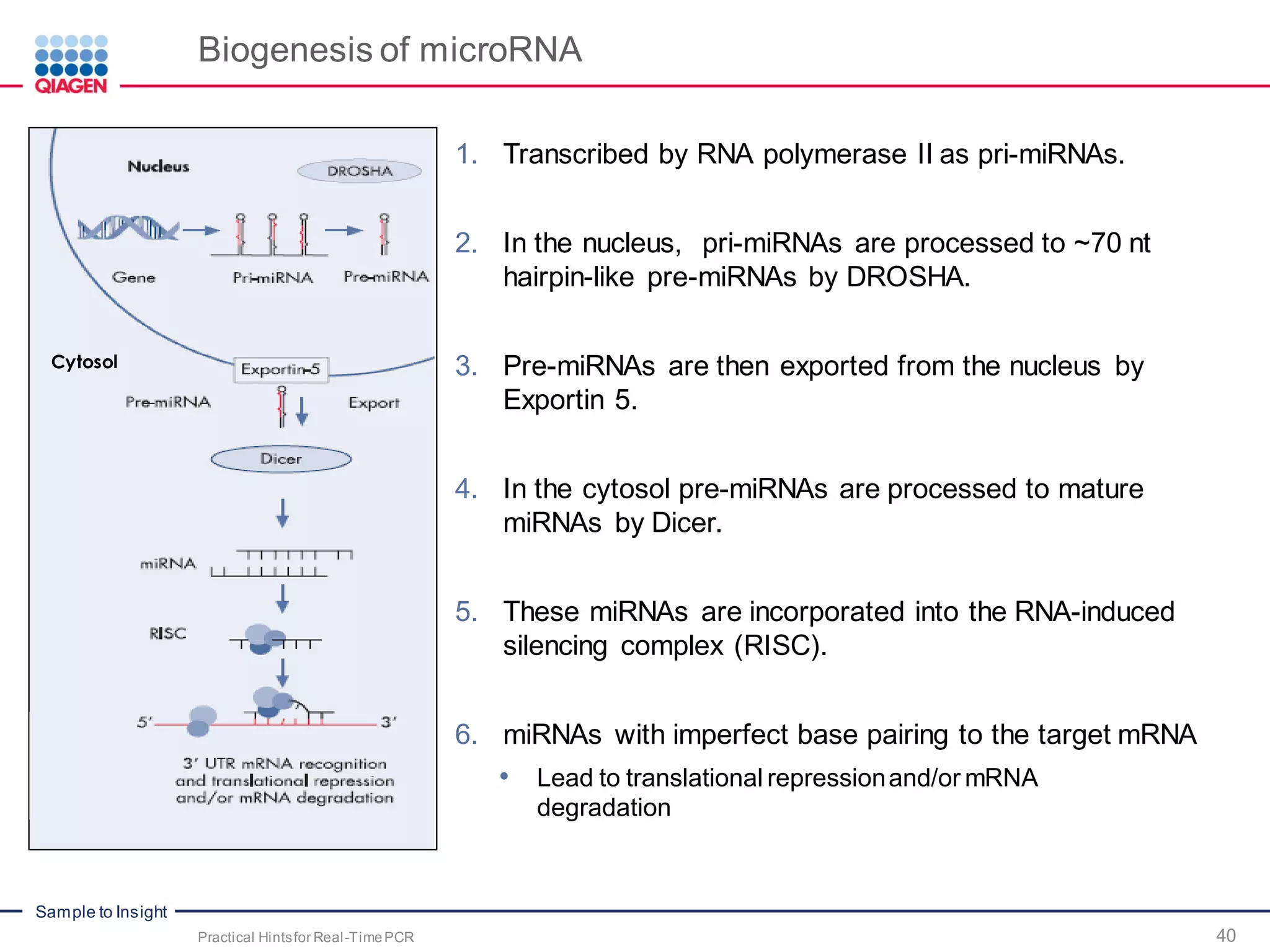
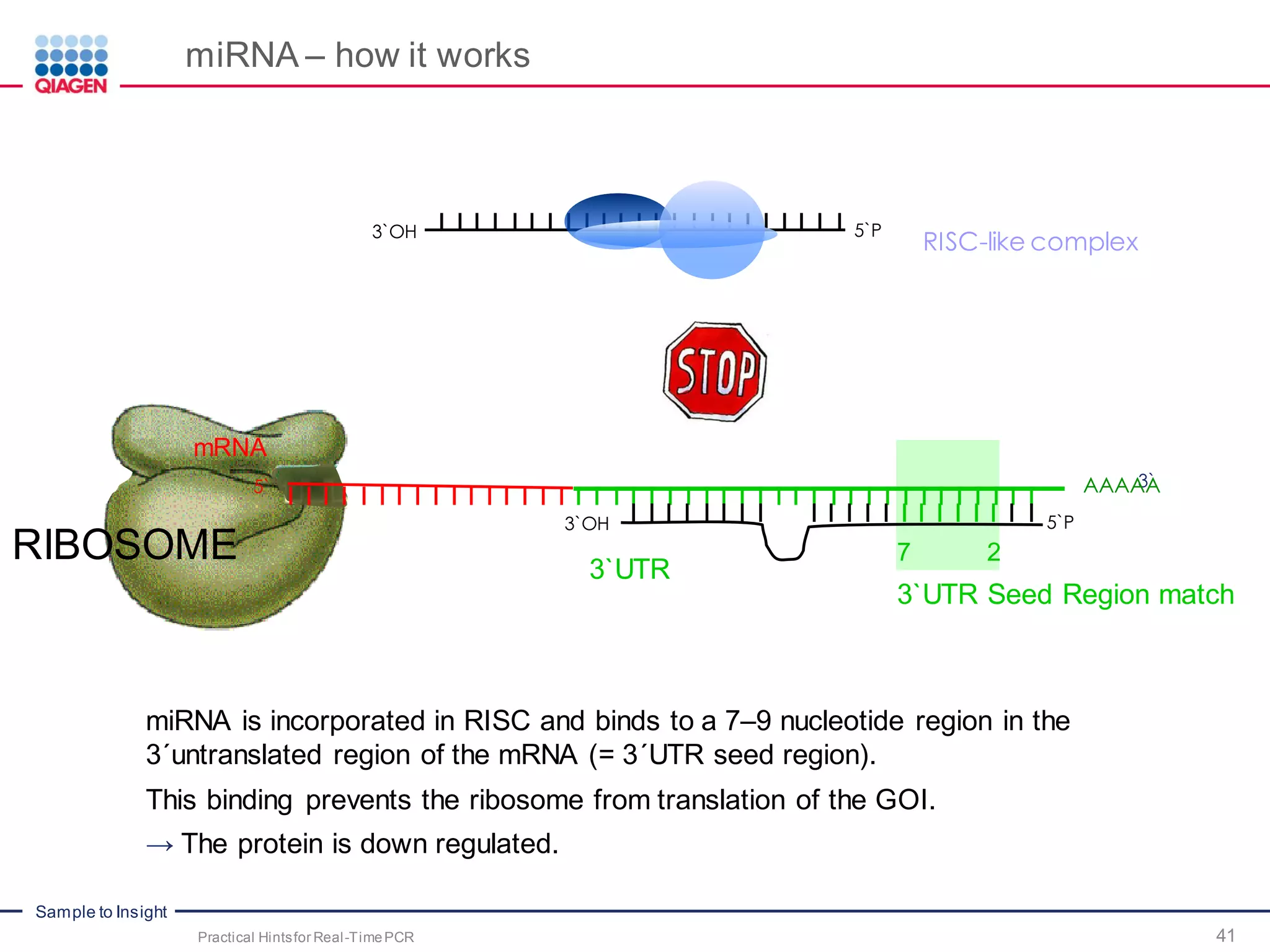
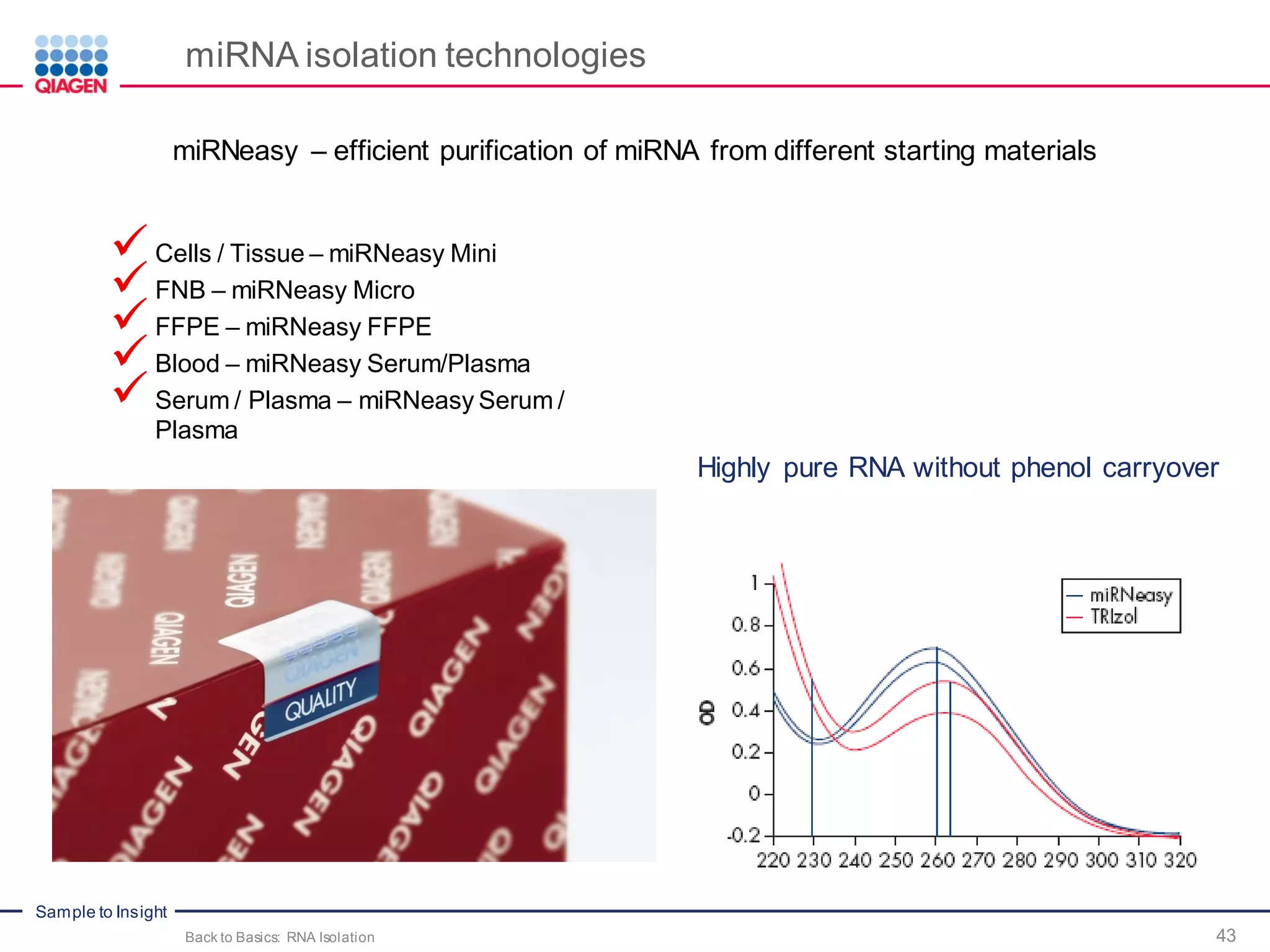
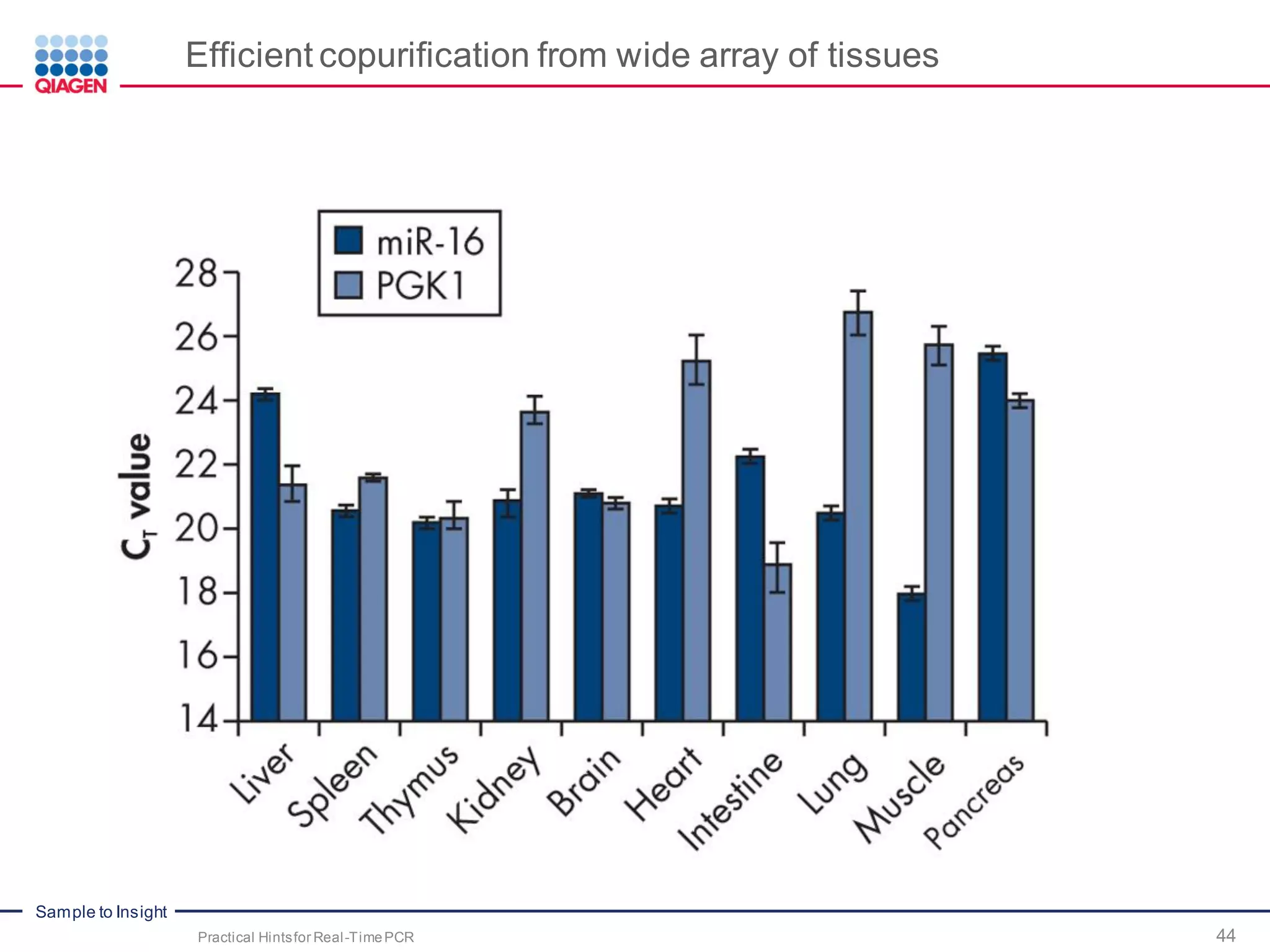
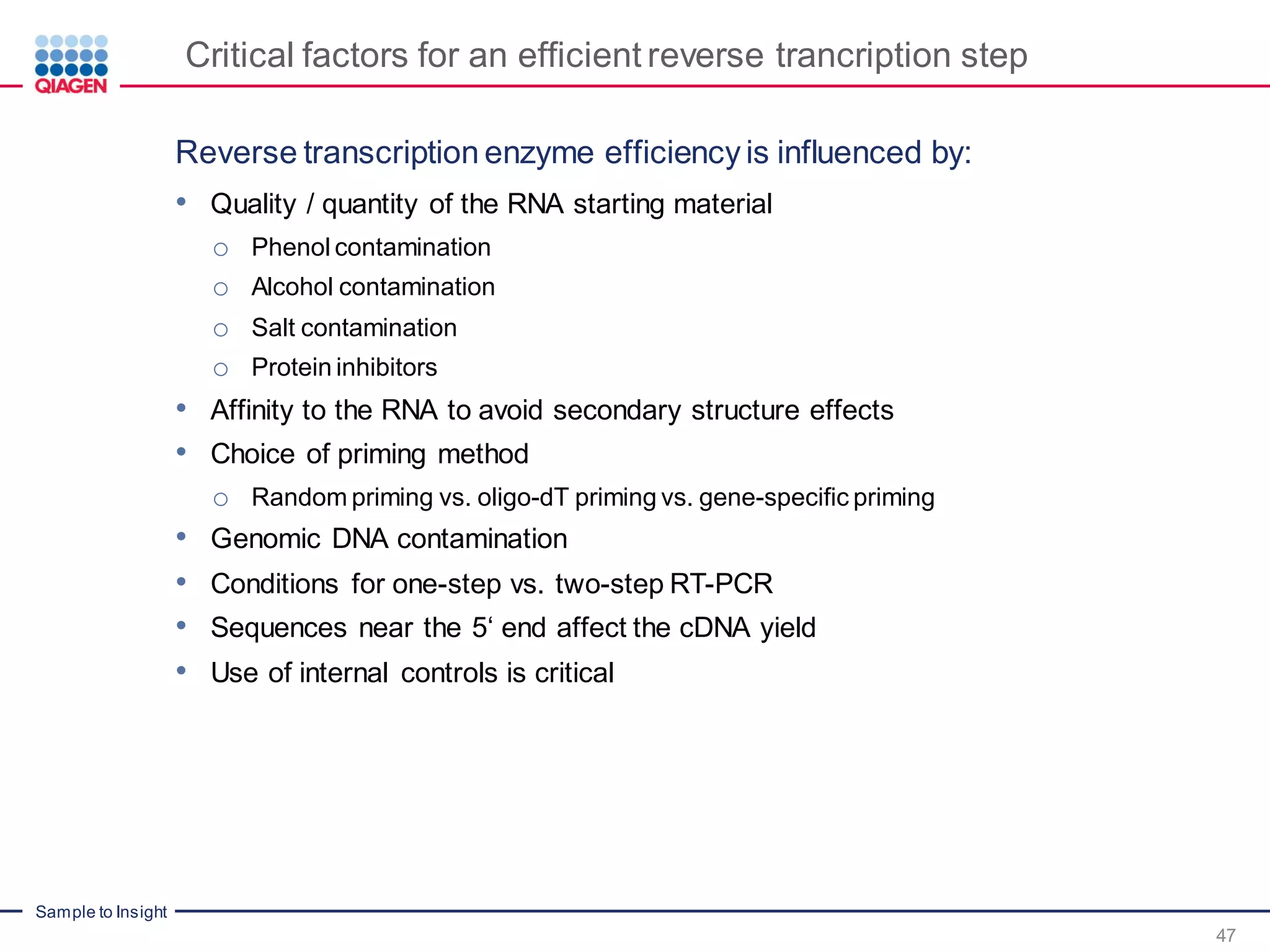
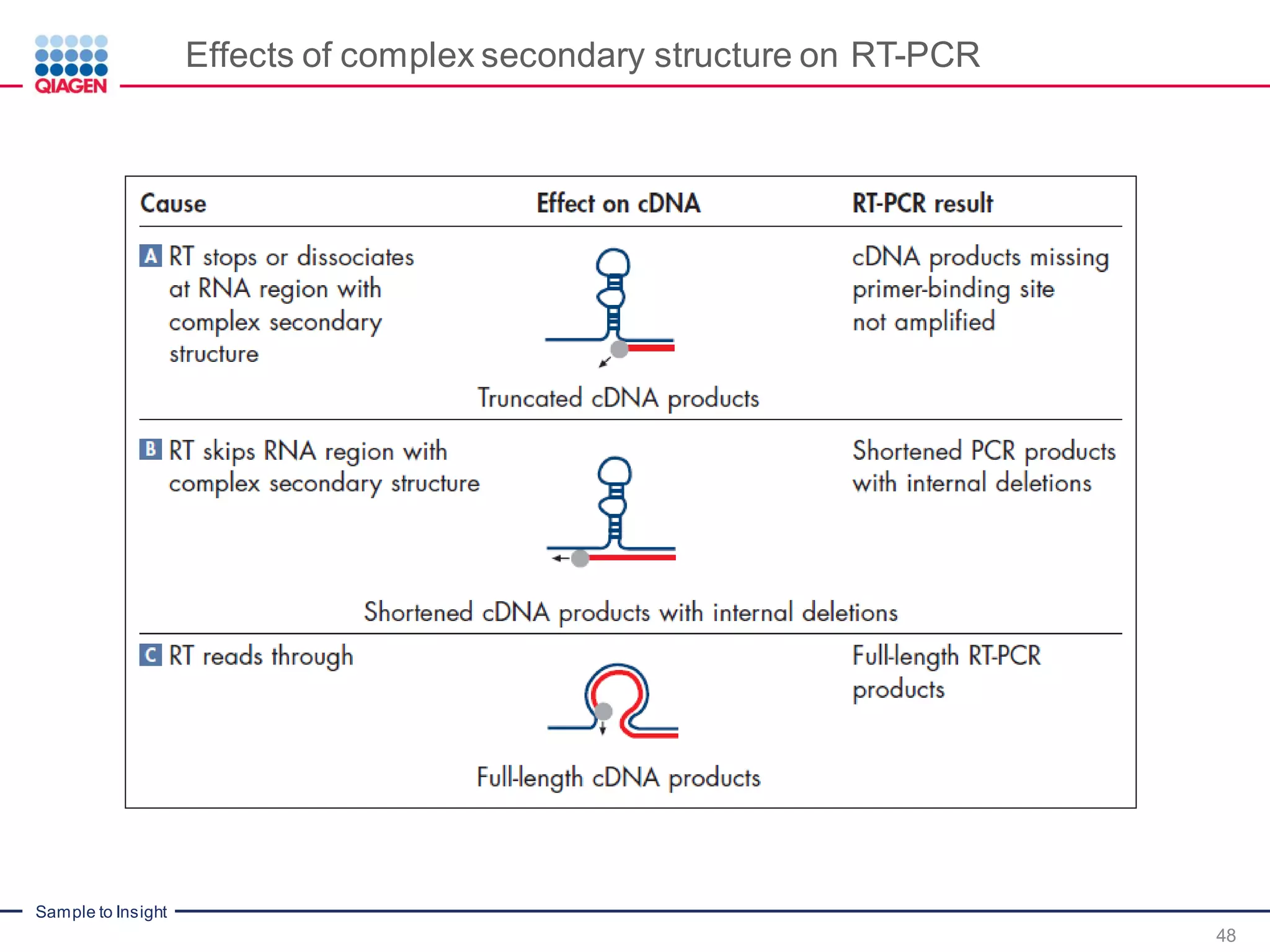
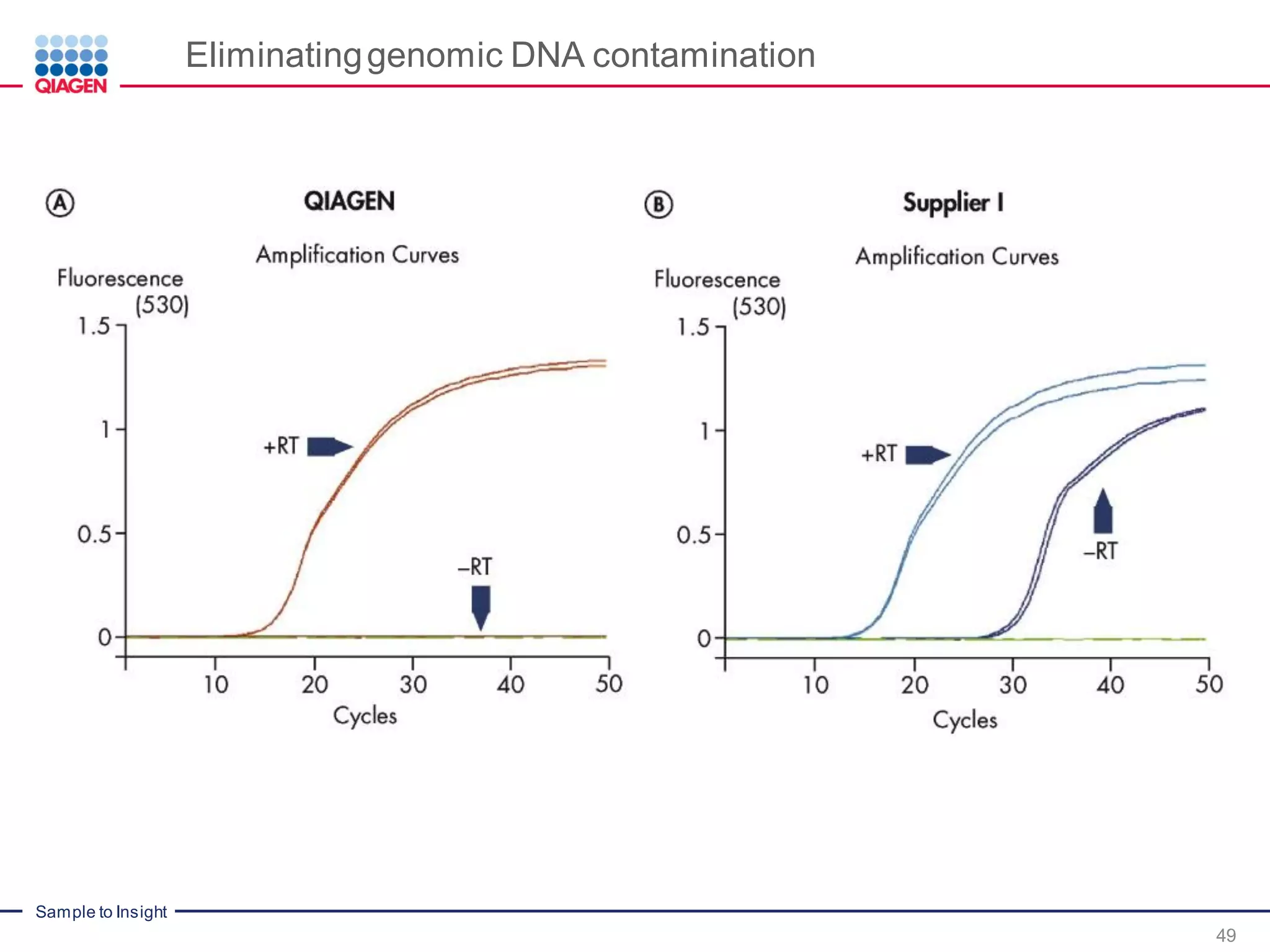
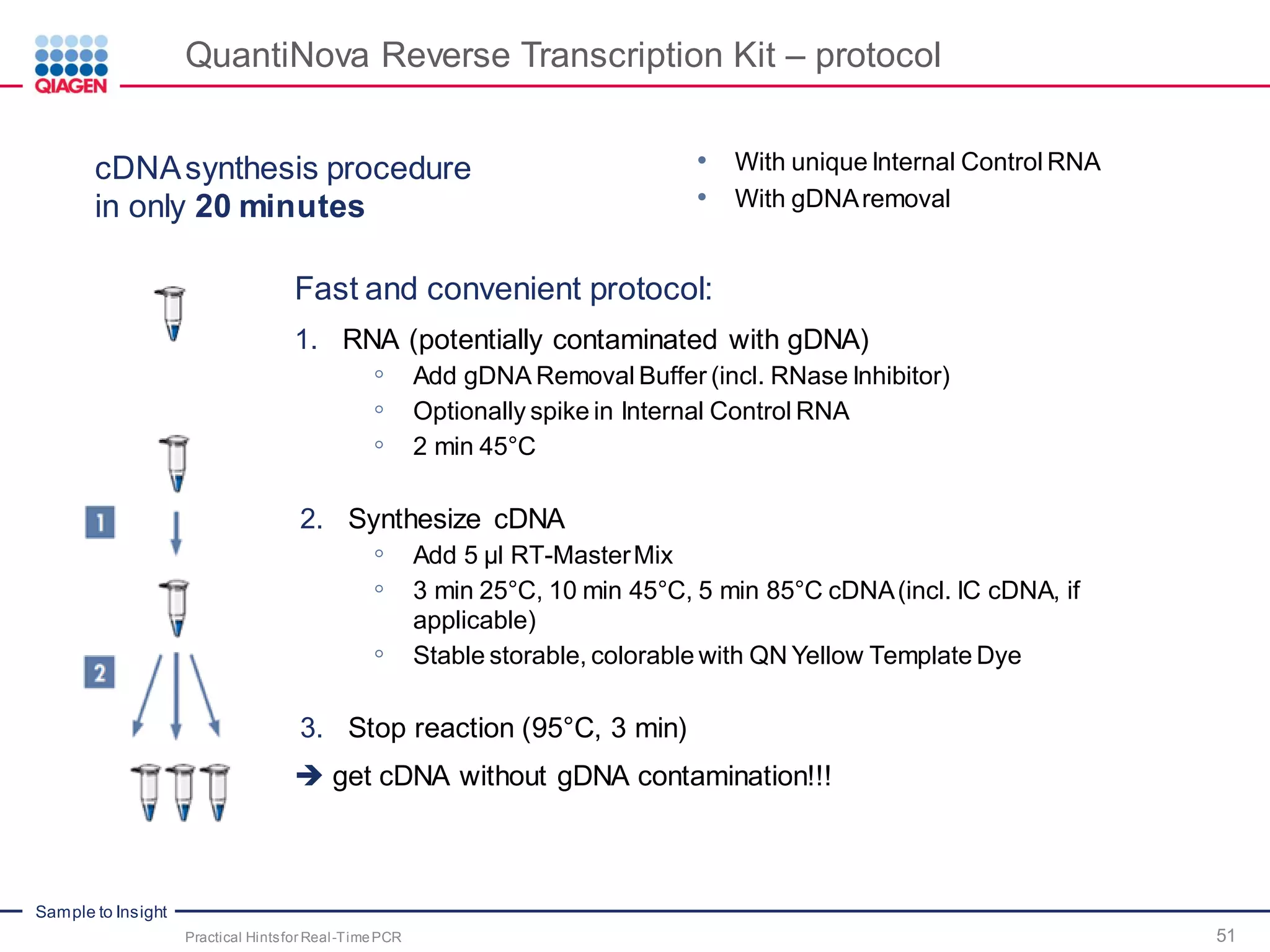
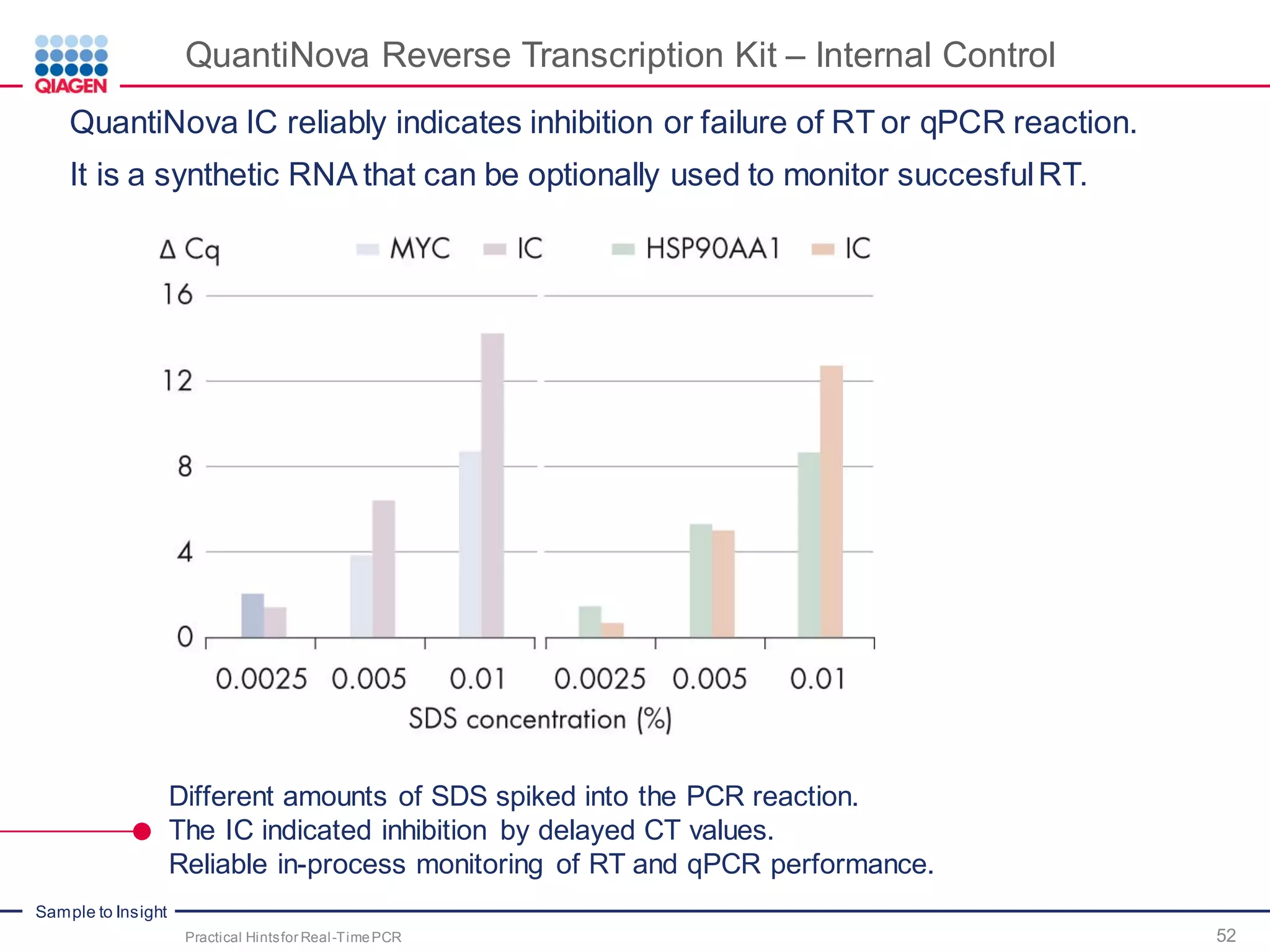
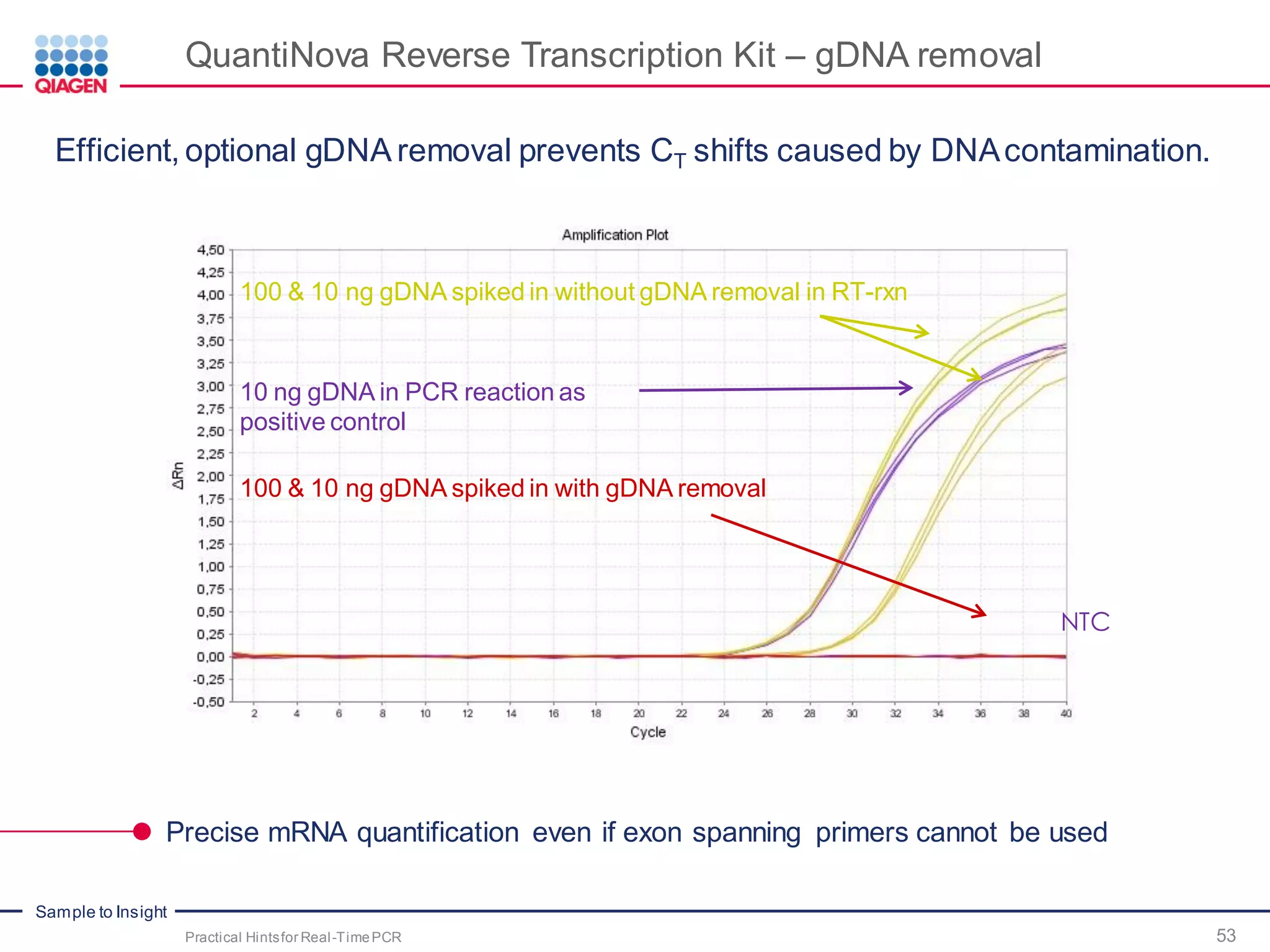
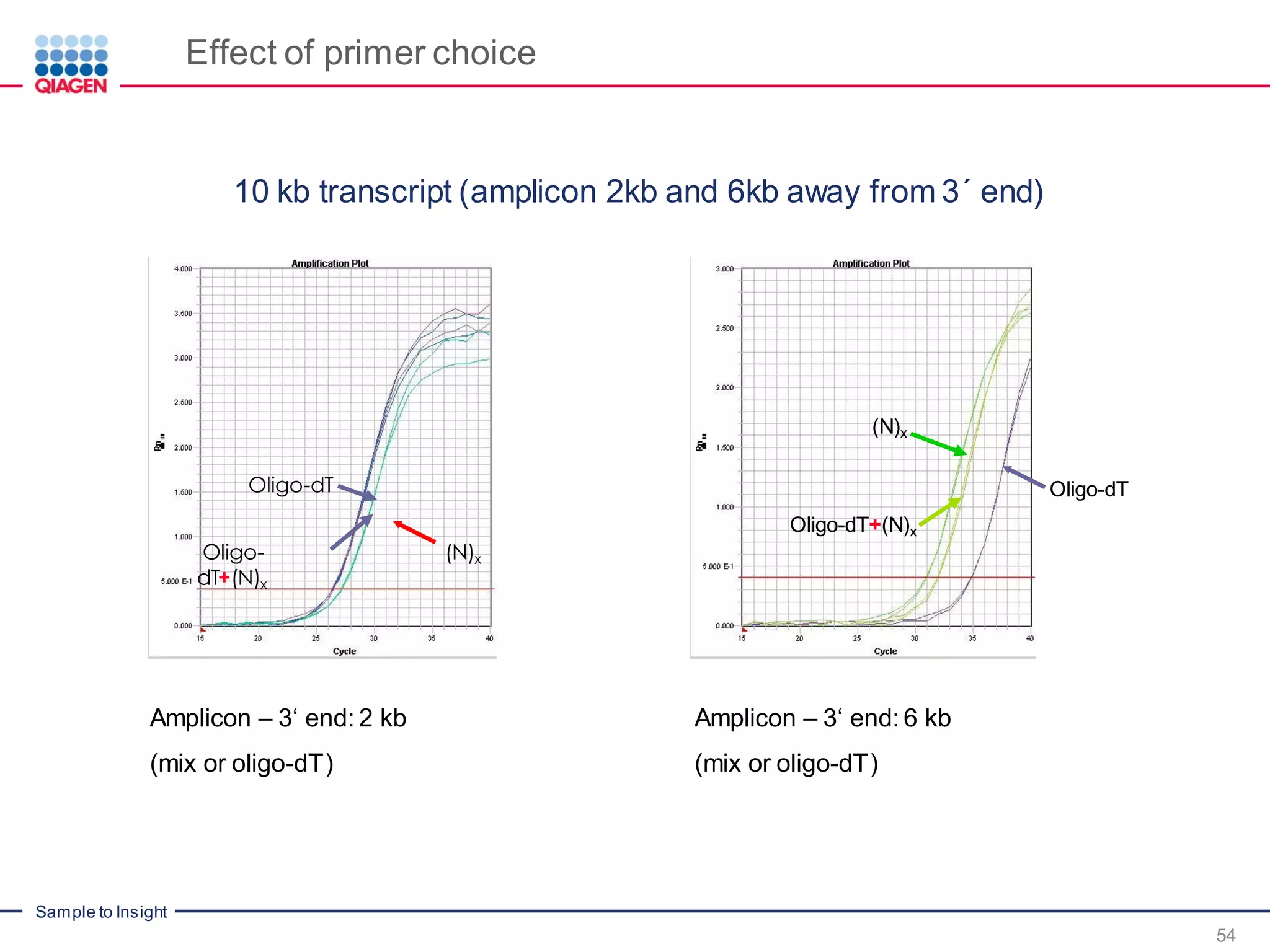
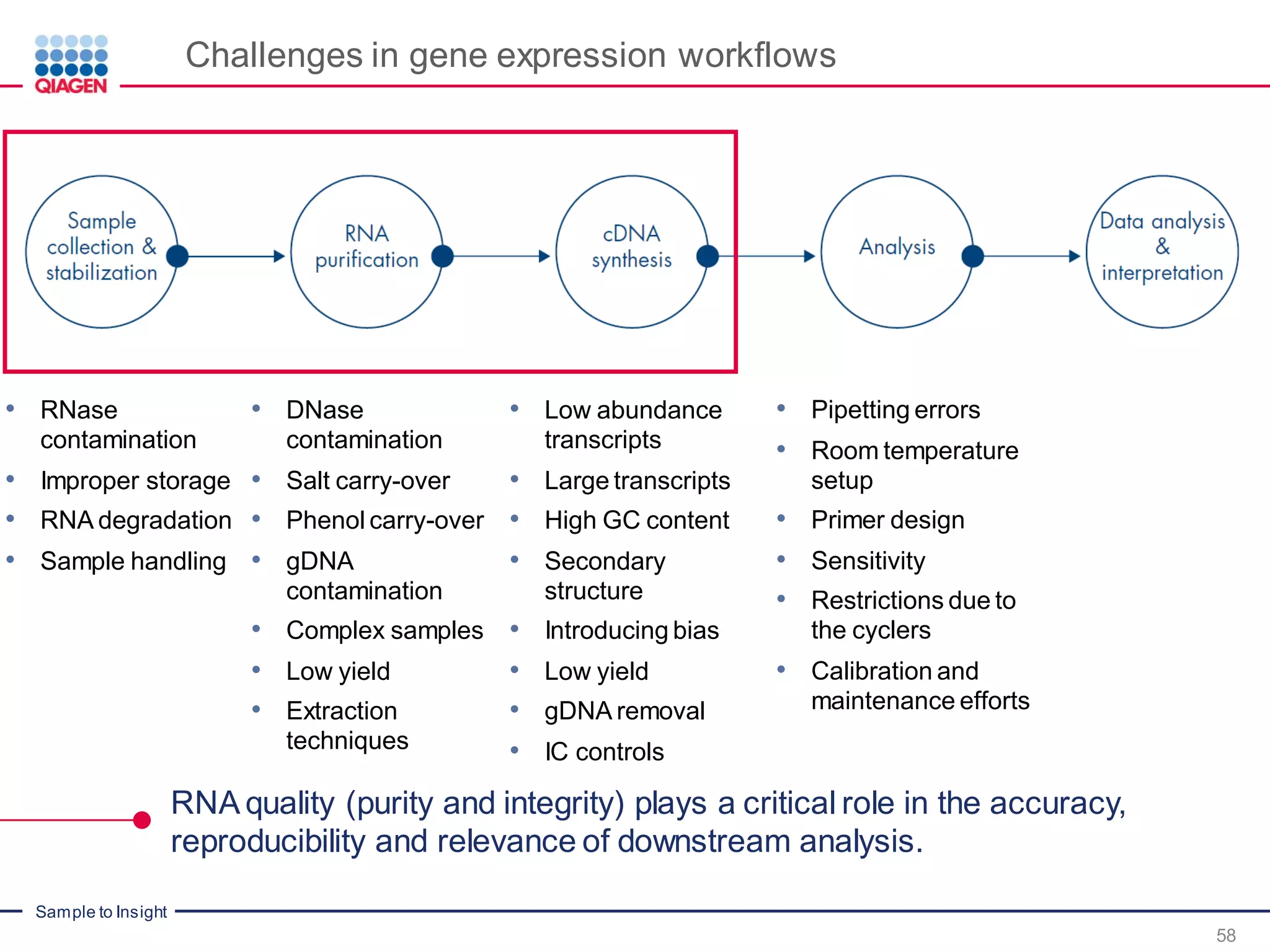
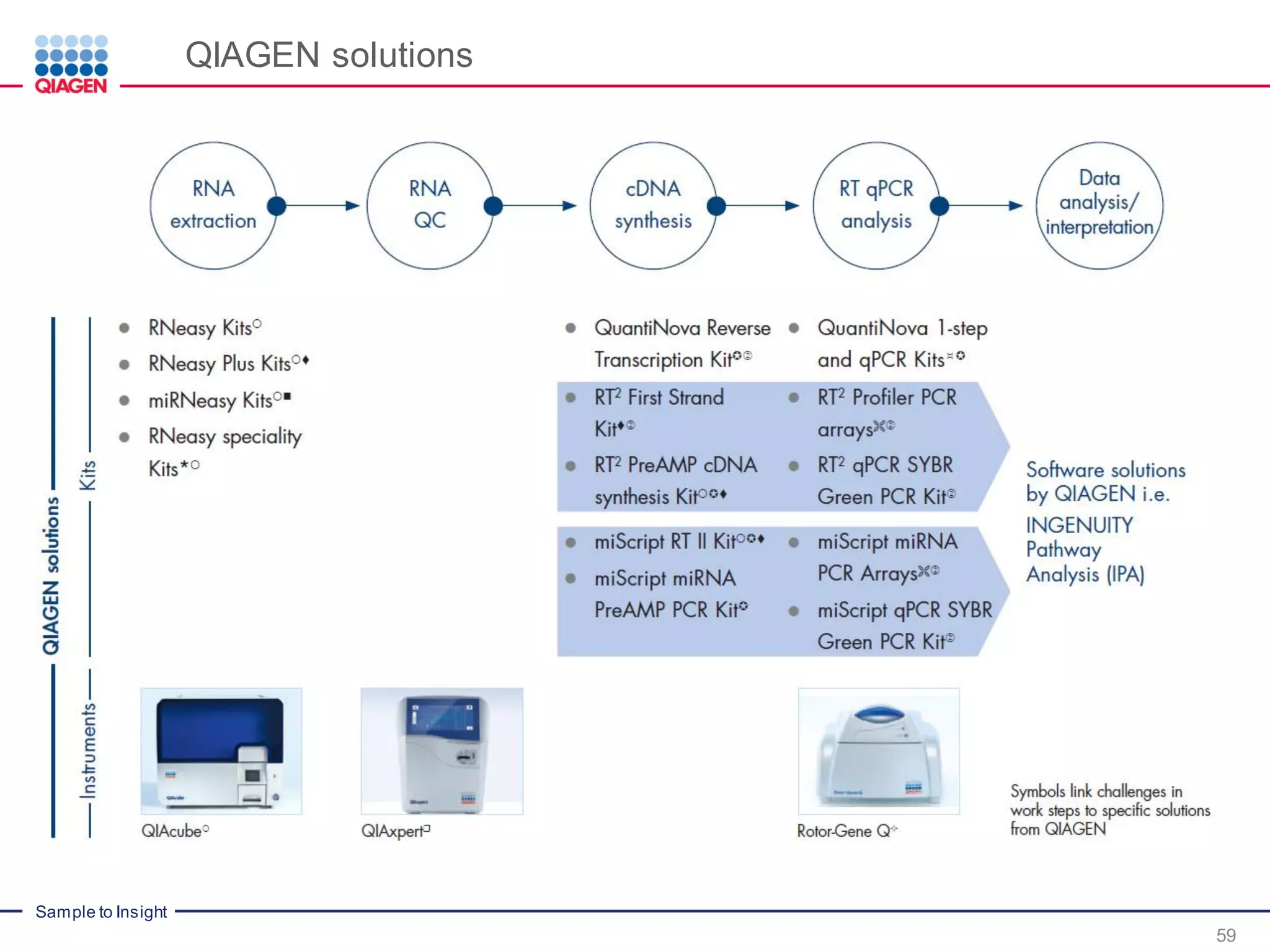
The document provides a comprehensive overview of critical steps for real-time PCR analysis and highlights the importance of RNA stabilization, isolation, and purification in achieving accurate gene expression results. It discusses various methods and challenges associated with gene expression workflows, including the removal of genomic contamination and specific considerations for different sample types like blood, tissue, and plant materials. Additionally, it addresses the significance of microRNAs in gene regulation and their potential applications in diagnostics and oncology.