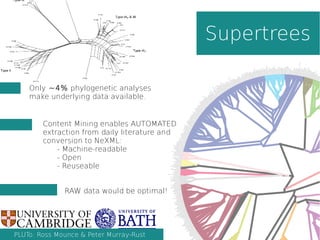
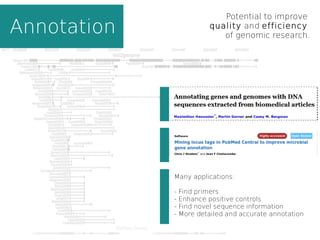
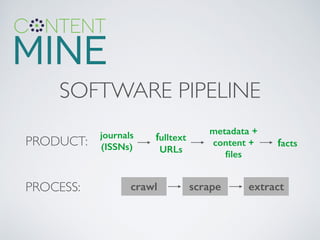
This document summarizes content mining technology and policy developments. It discusses what content and mining are, provides a brief history of content mining, and outlines legal considerations around copyright and database rights. It then describes the ContentMine software and pipeline for scraping, normalizing, and extracting facts from scholarly documents at scale. Examples of mining applications in chemistry, clinical trials, phylogenetics, and genome annotation are provided. The document concludes with a discussion of the potential value of content mining for public health researchers.











![BASIC SCRAPER JSON
name of the scraper:
the URL(s) it applies to:
the elements to capture:
element name:
where to find it:
{!
"name": "PLOS",!
"url": "plosw*.org",!
"elements": {!
"title": {!
"selector": “//h1[@property=‘dc:title’]”,!
}!
}!
}!
http://github.com/ContentMine/scraperJSON](https://image.slidesharecdn.com/who-slides-full-150511073736-lva1-app6892/85/ContentMine-Presentation-for-WHO-Health-Data-Seminar-14-320.jpg)

![SCRAPERS
{!
"name": "PLoS",!
"url": "plosw*.org",!
"elements": {!
"title": {!
"selector": “//h1[@property=‘dc:title’]”,!
}!
}!
}!](https://image.slidesharecdn.com/who-slides-full-150511073736-lva1-app6892/85/ContentMine-Presentation-for-WHO-Health-Data-Seminar-16-320.jpg)