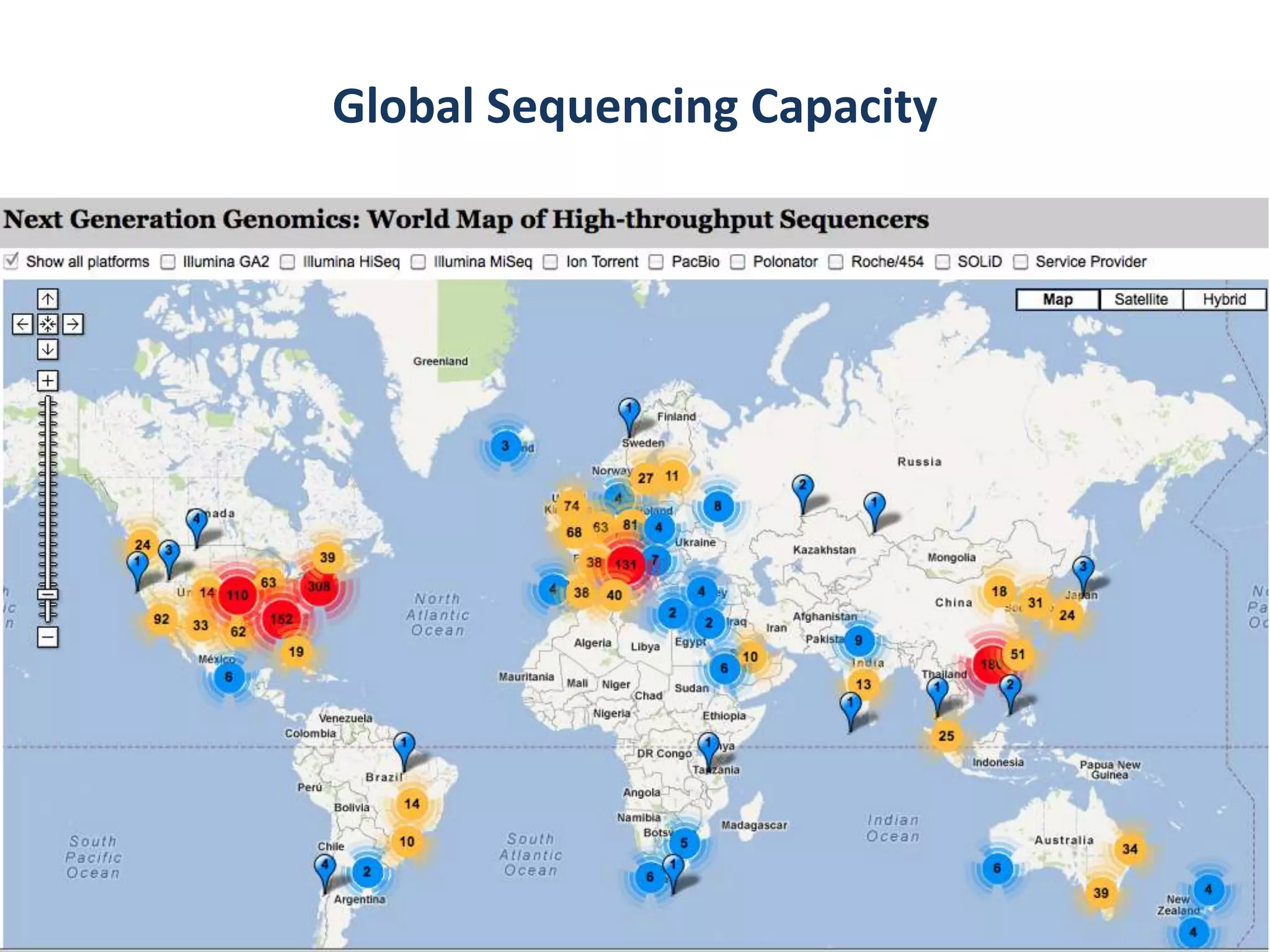
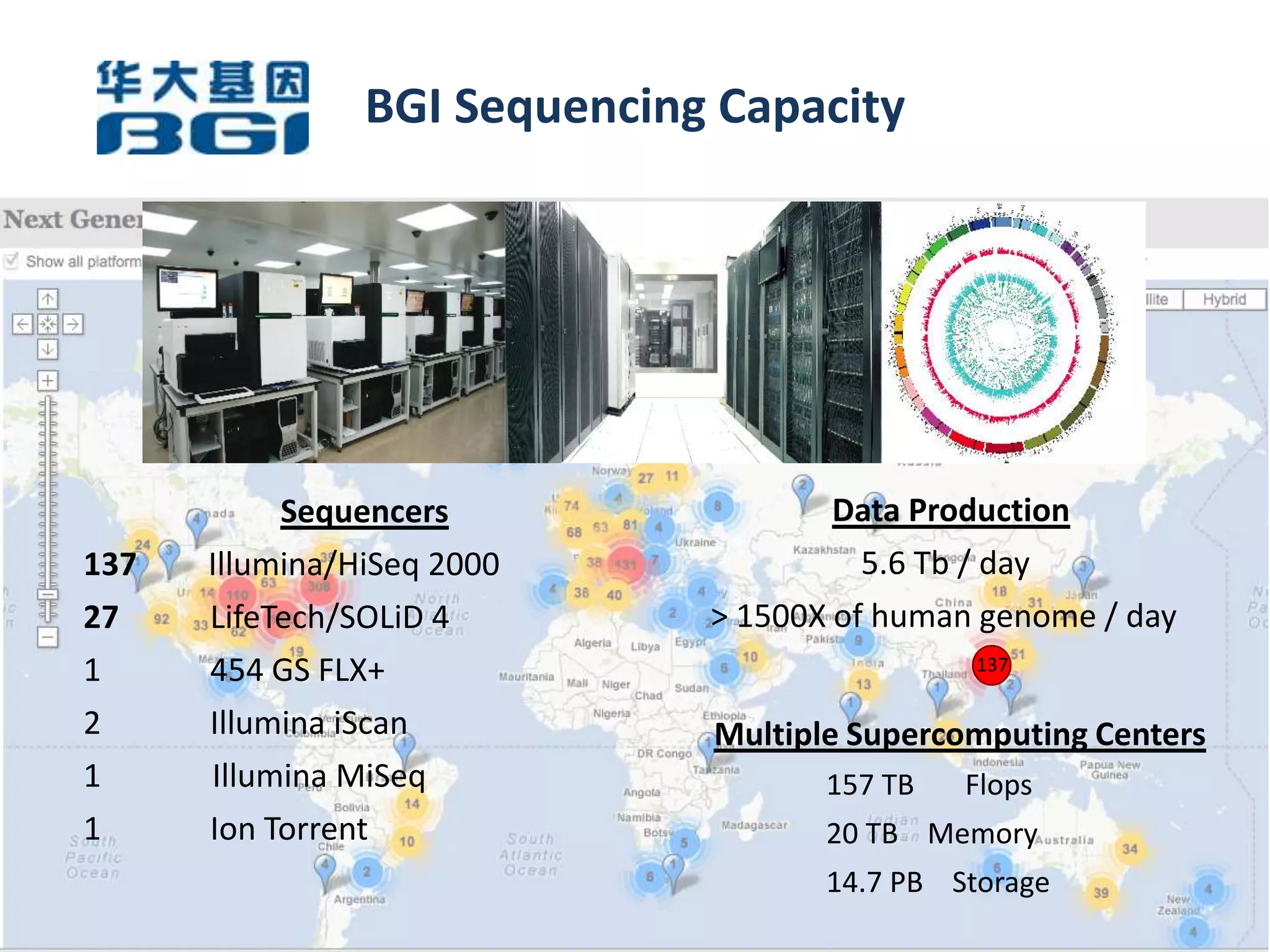
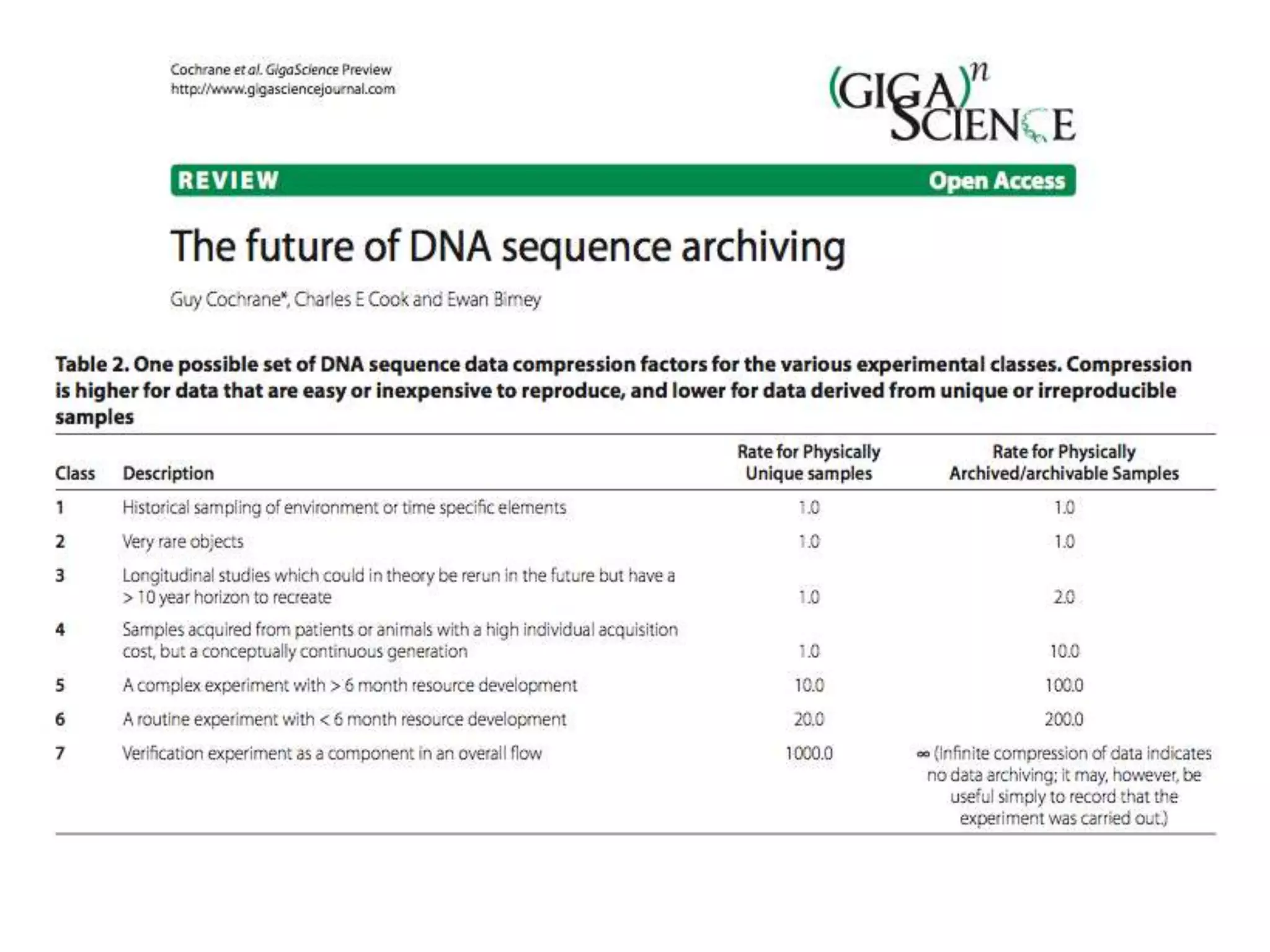
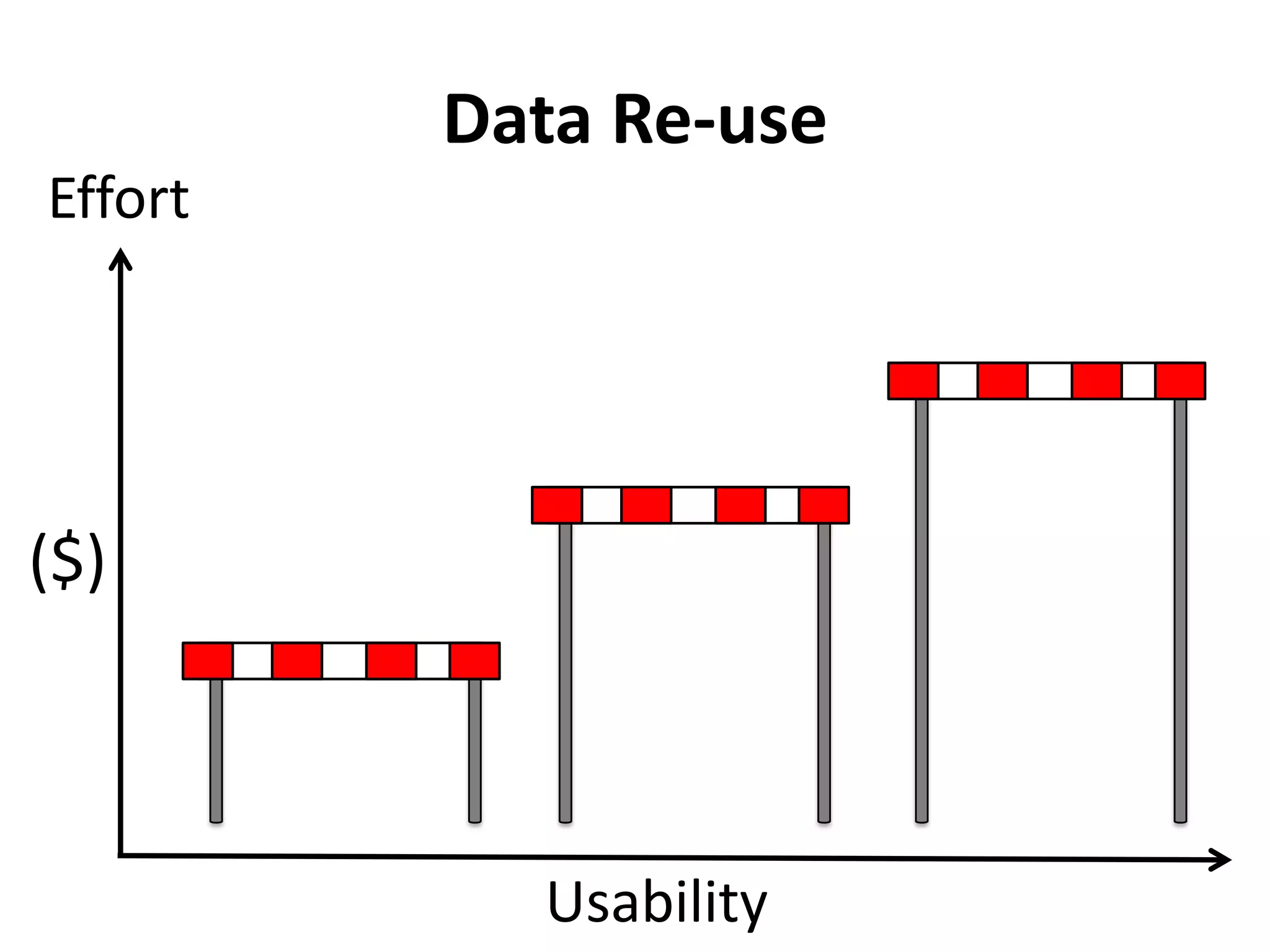
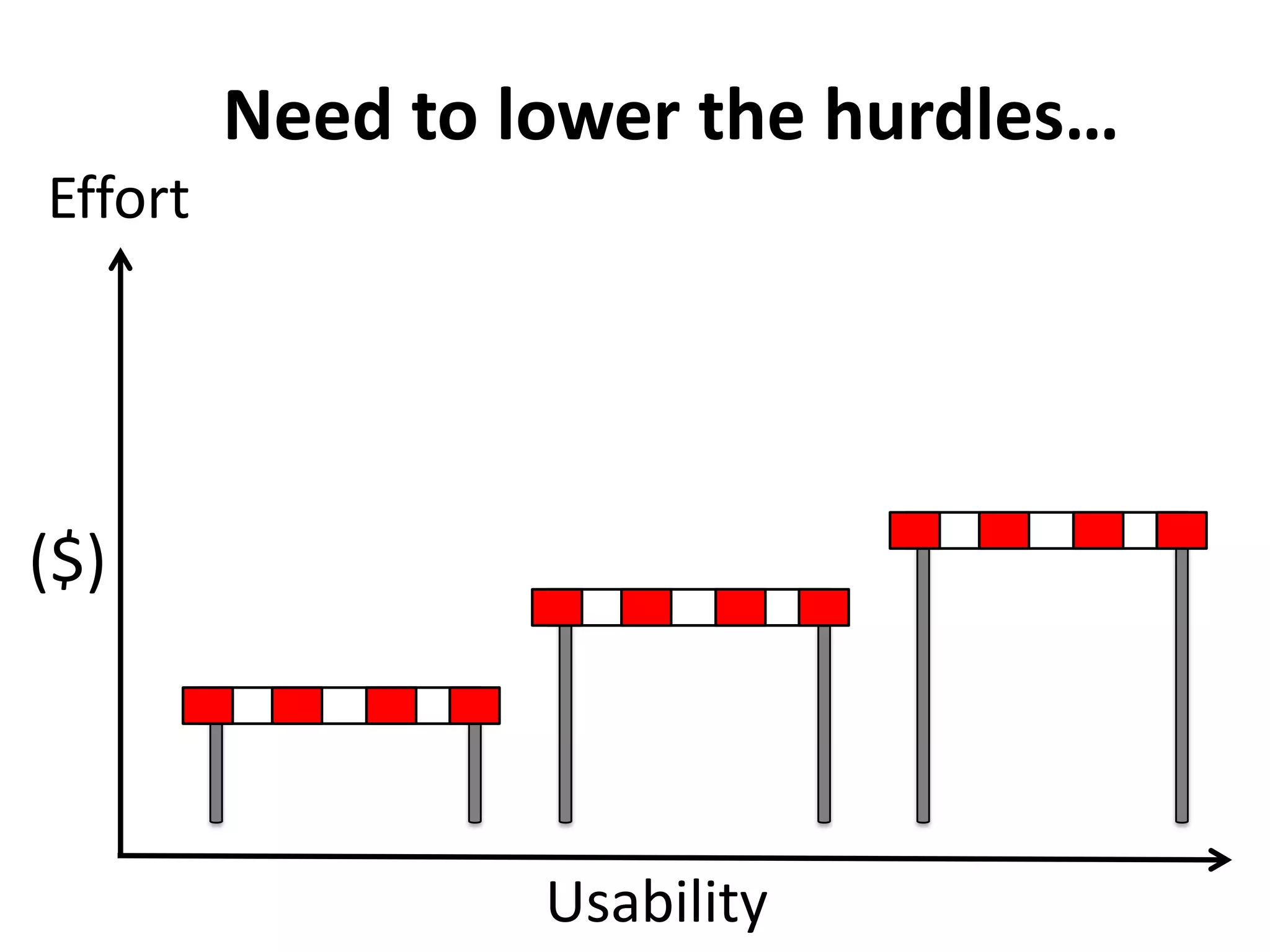
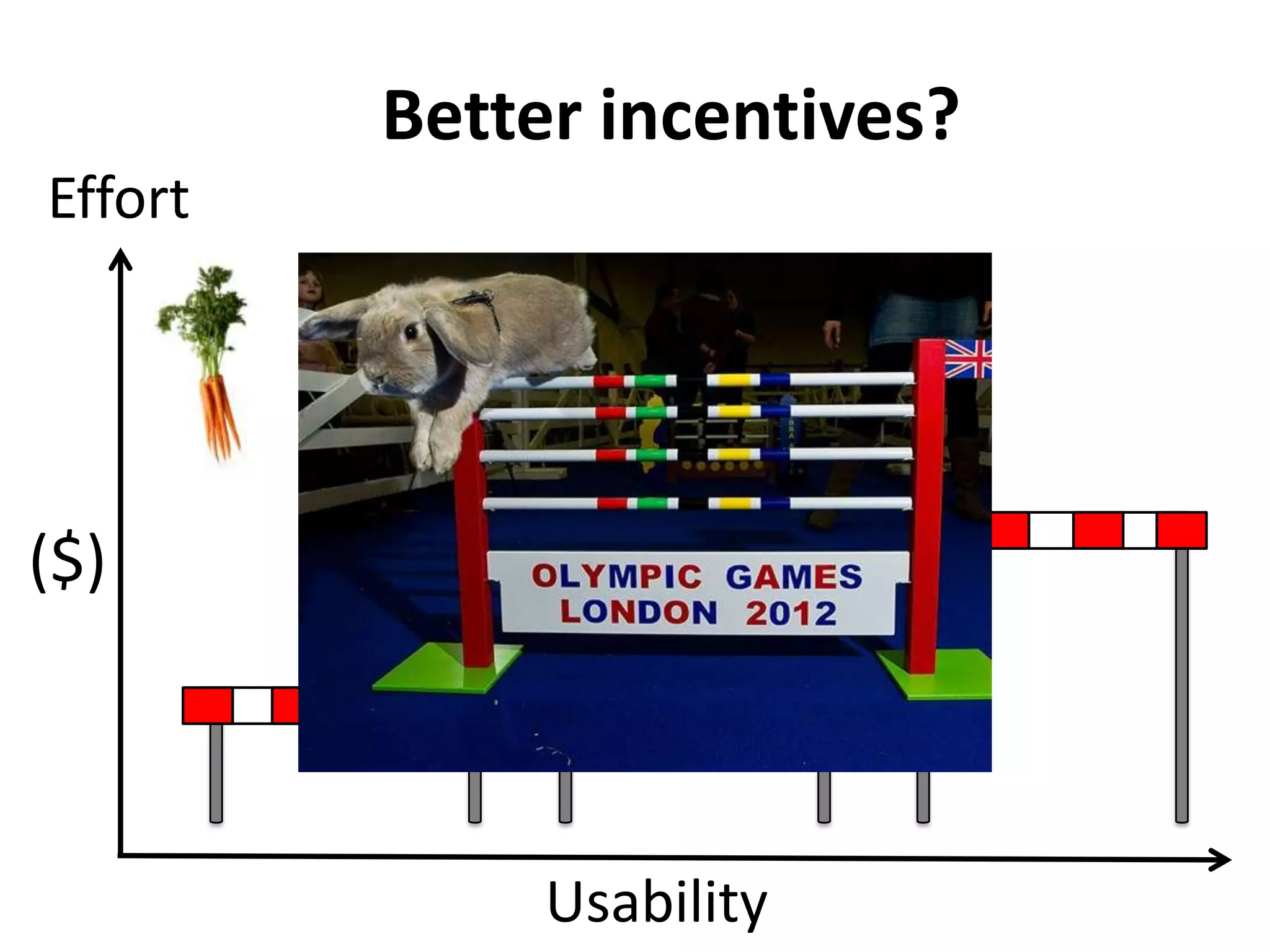
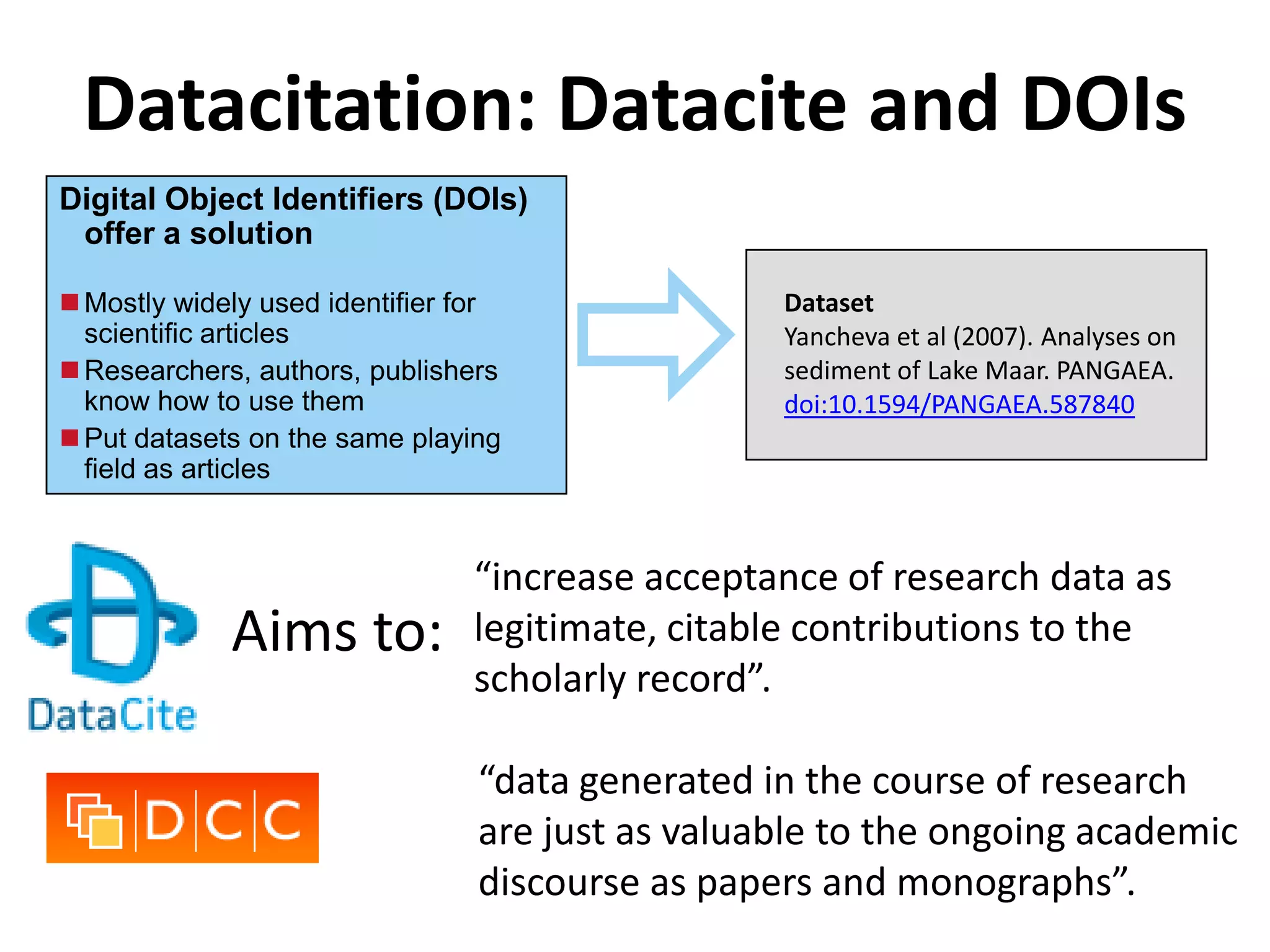
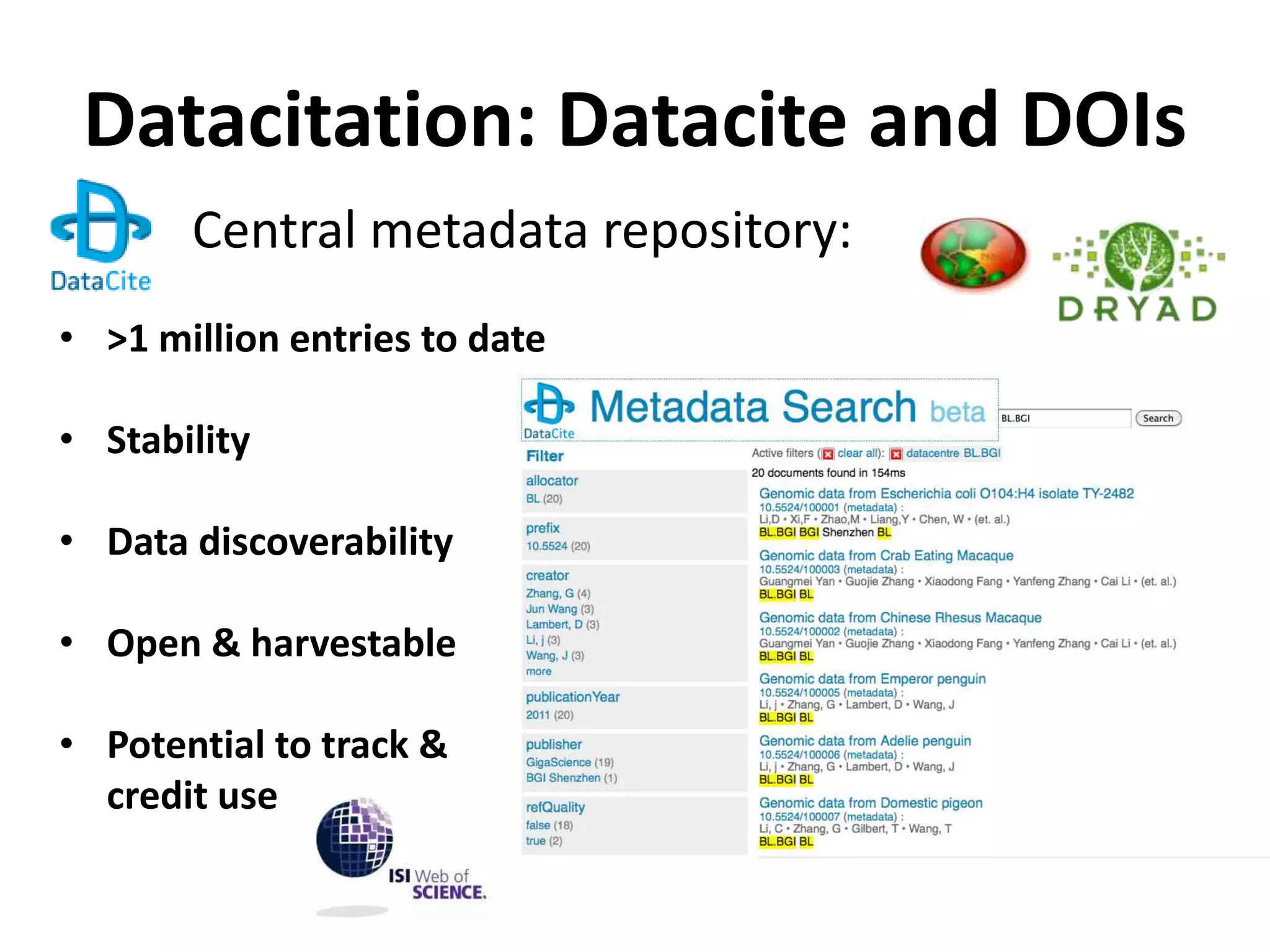
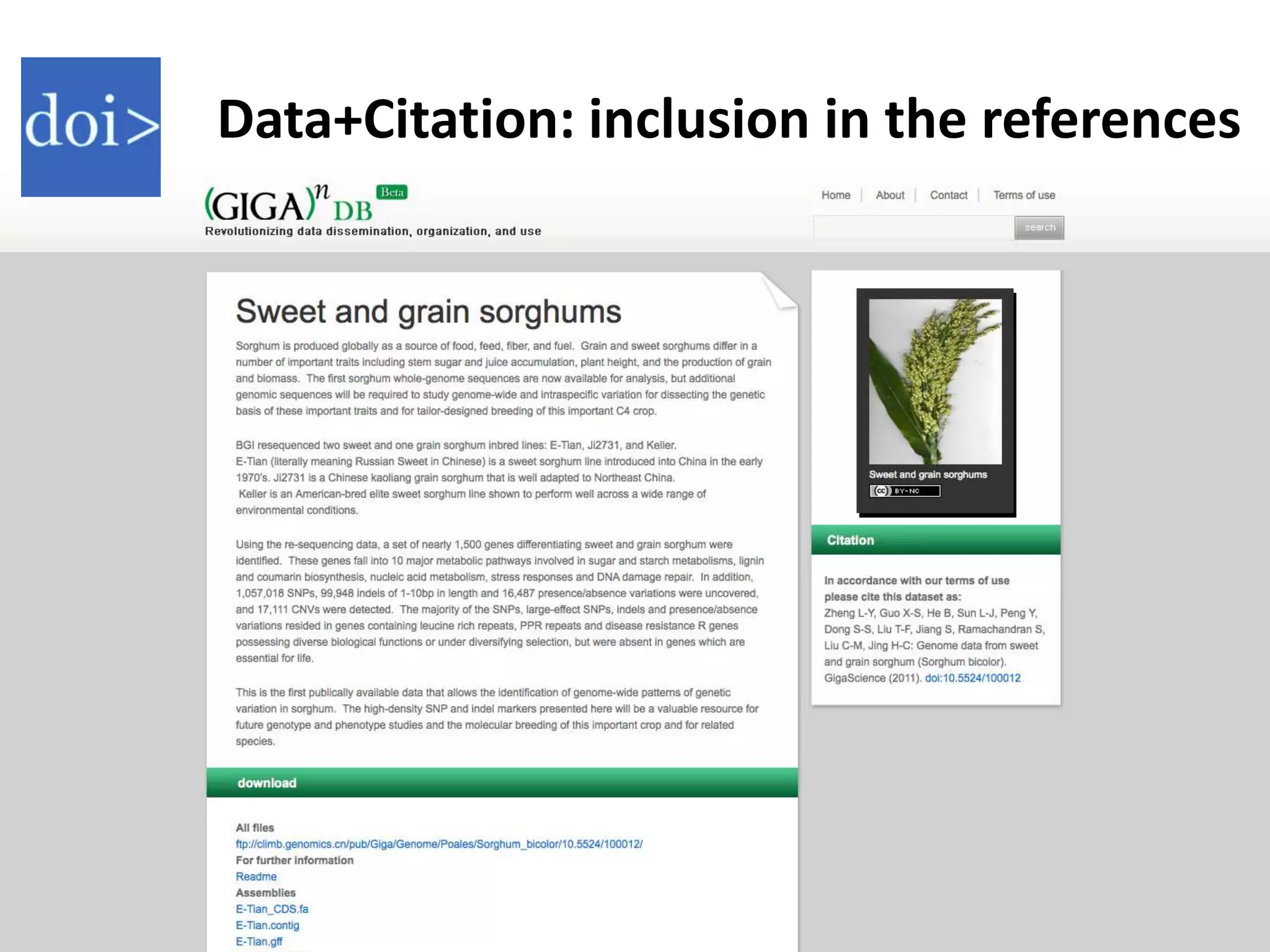
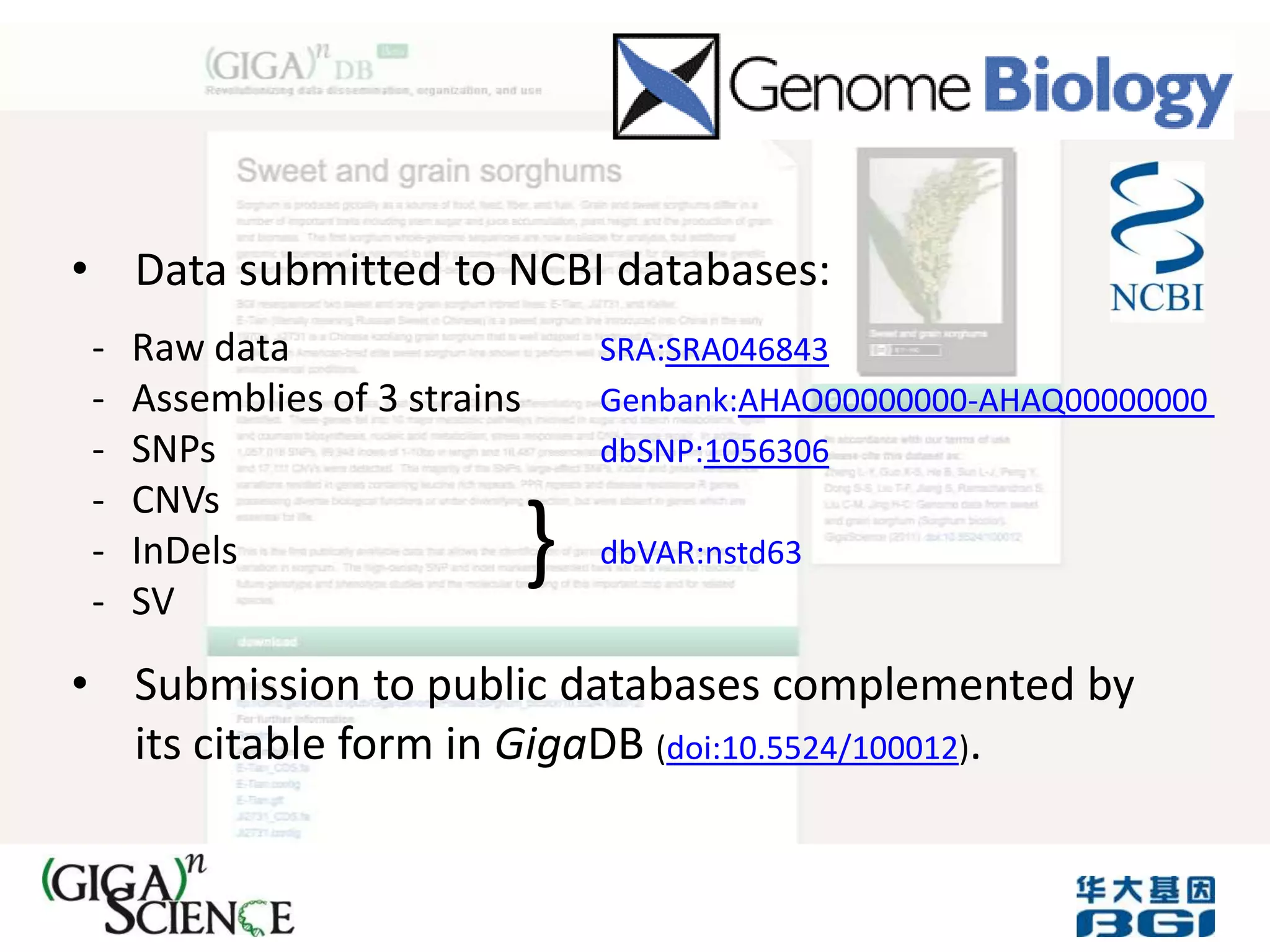
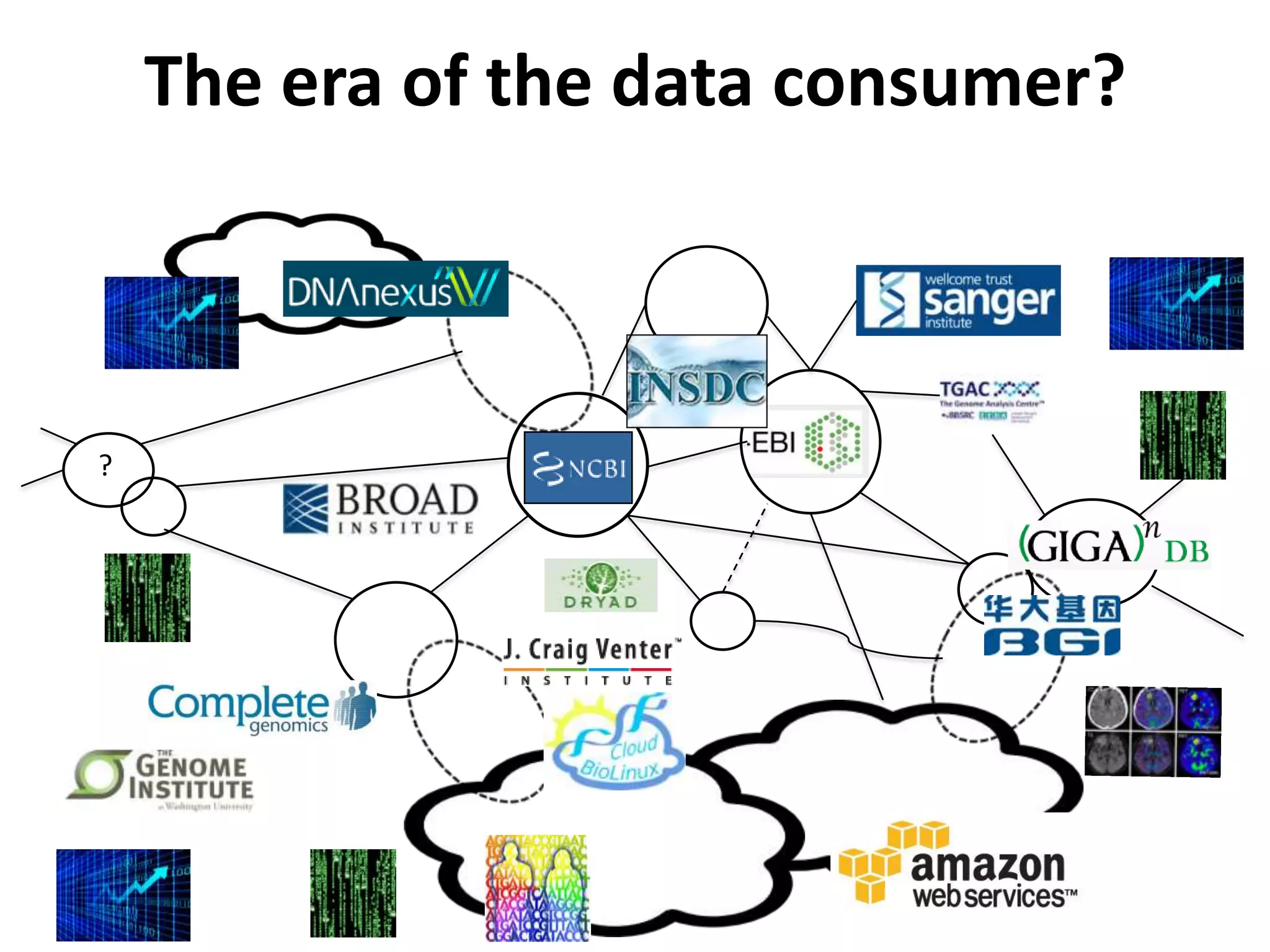
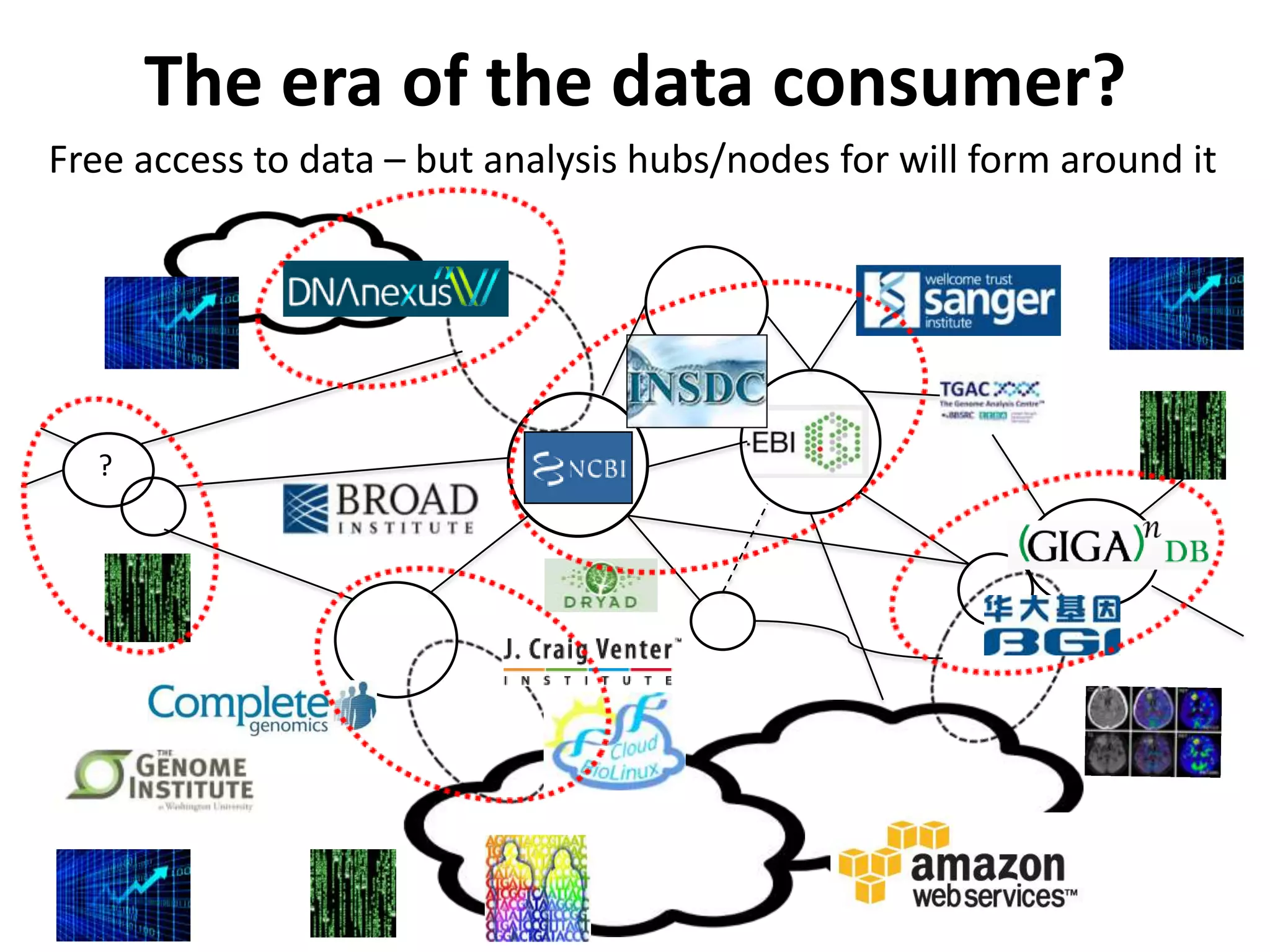
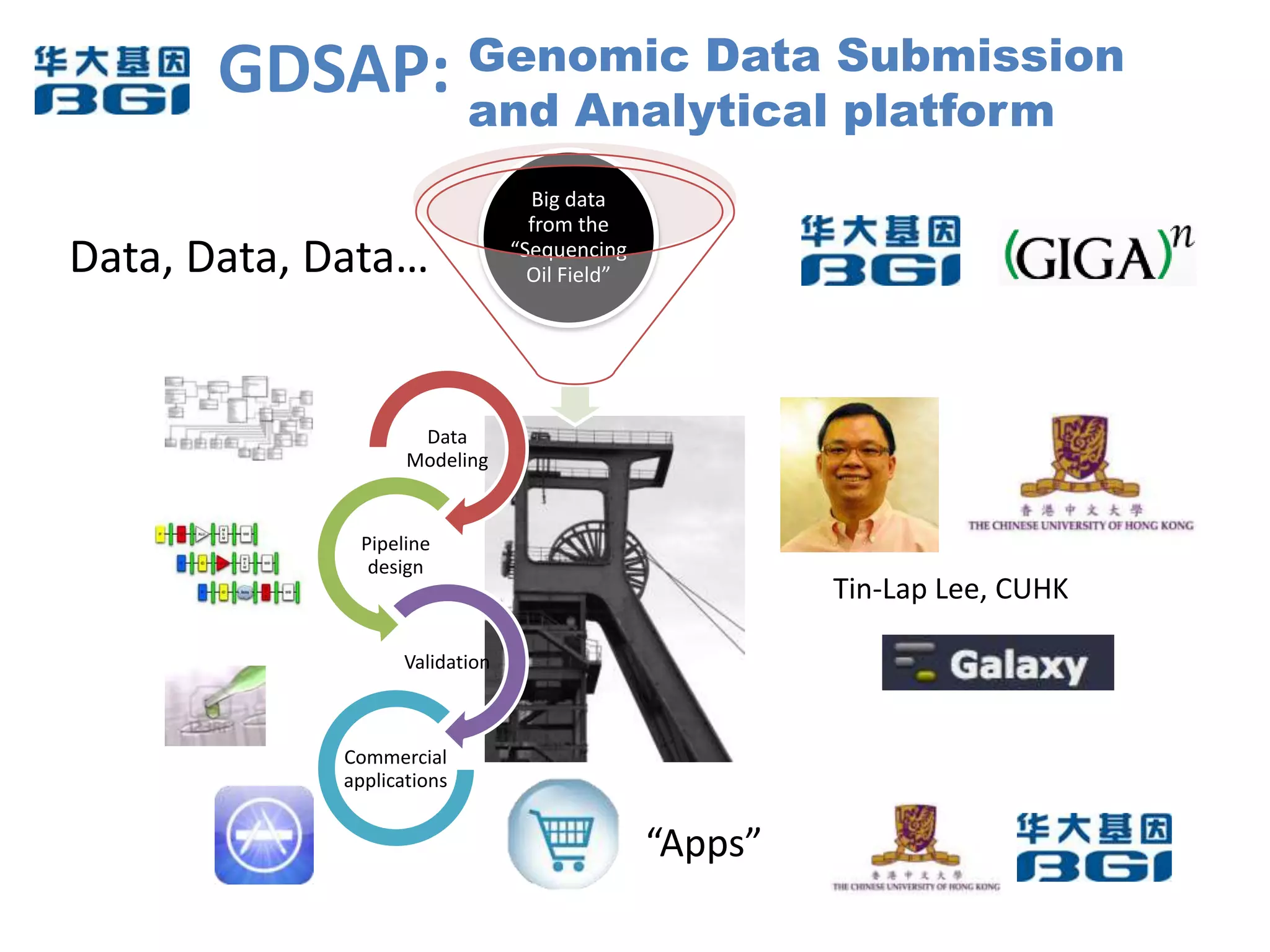
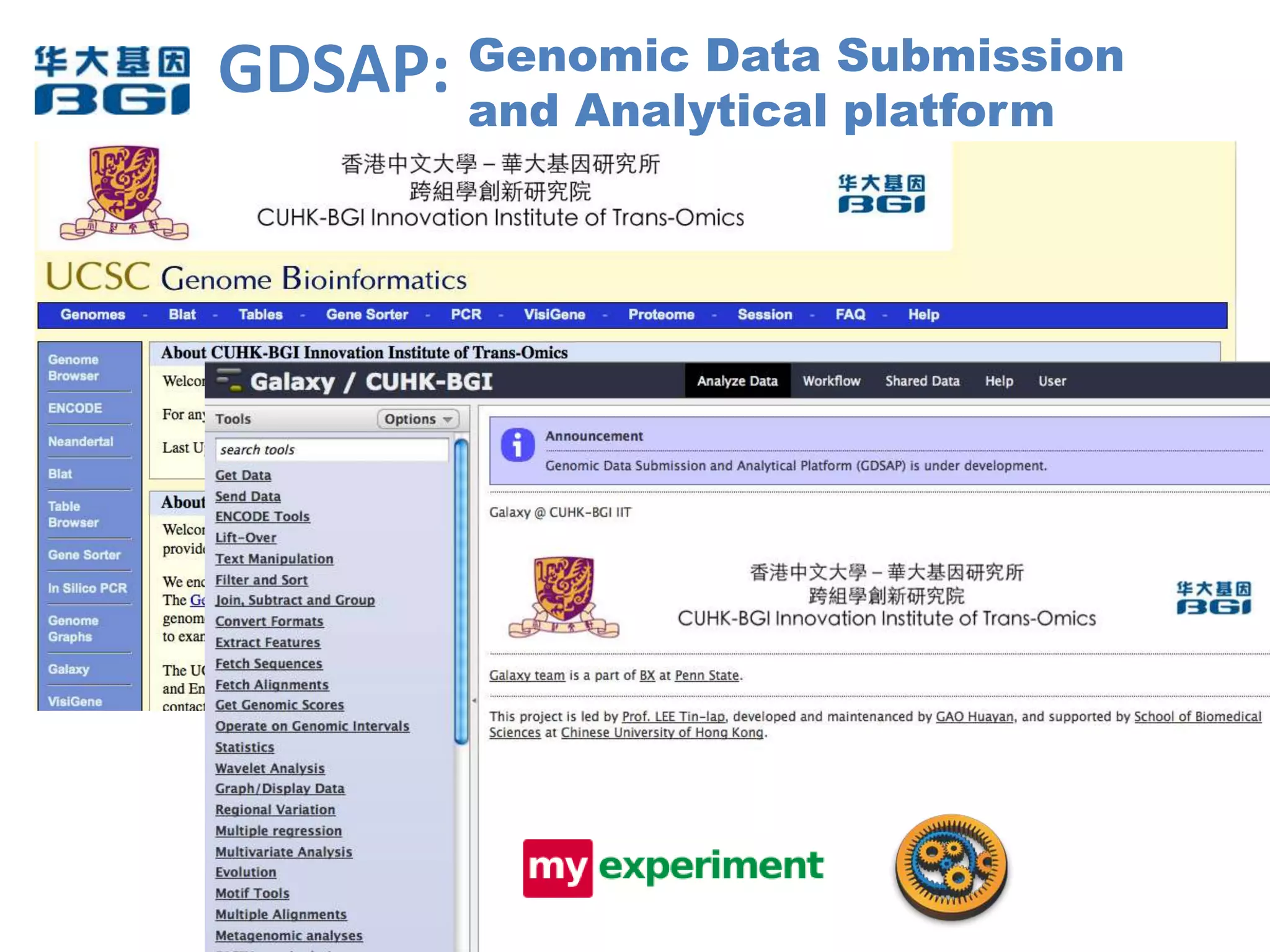
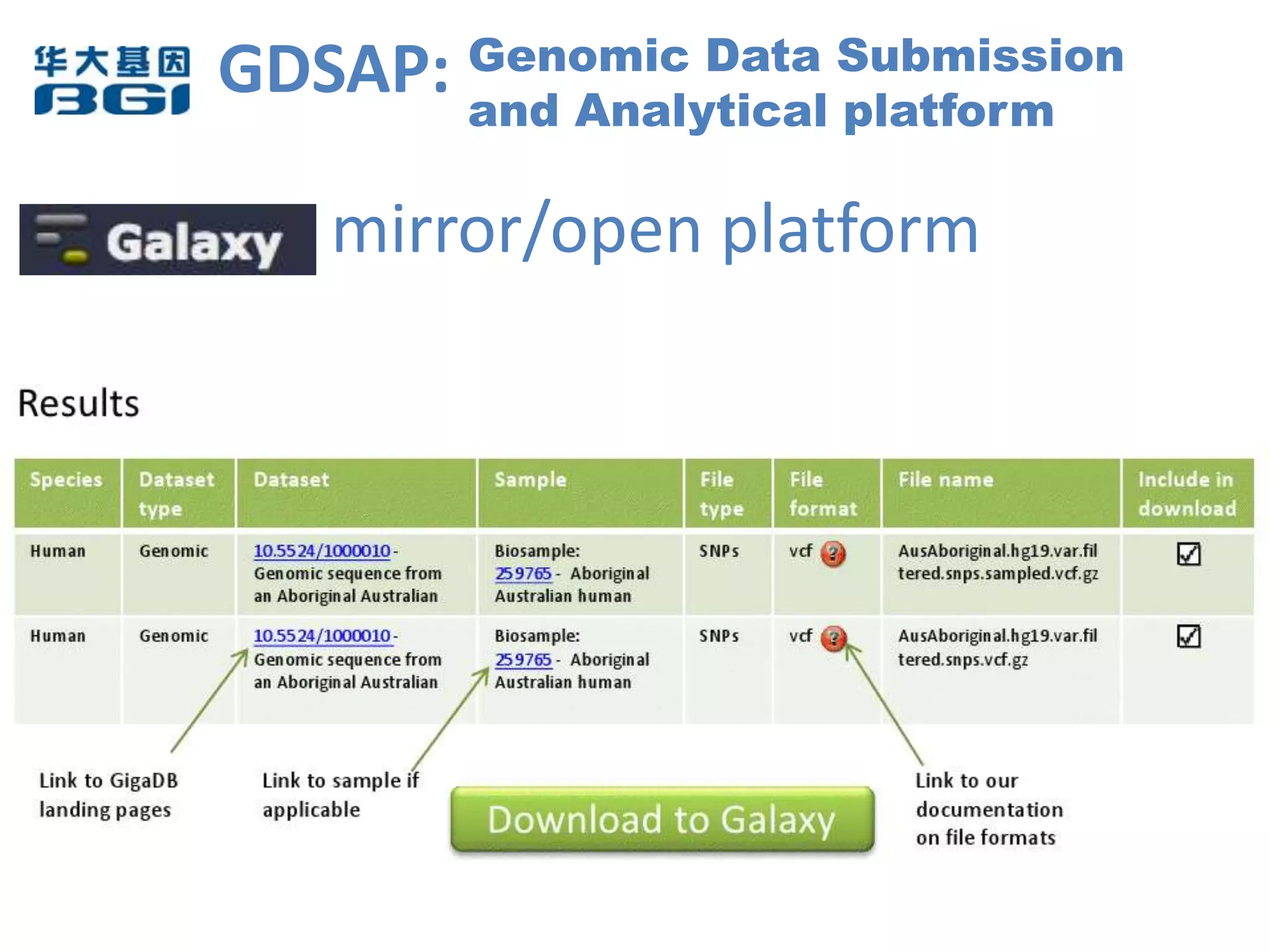
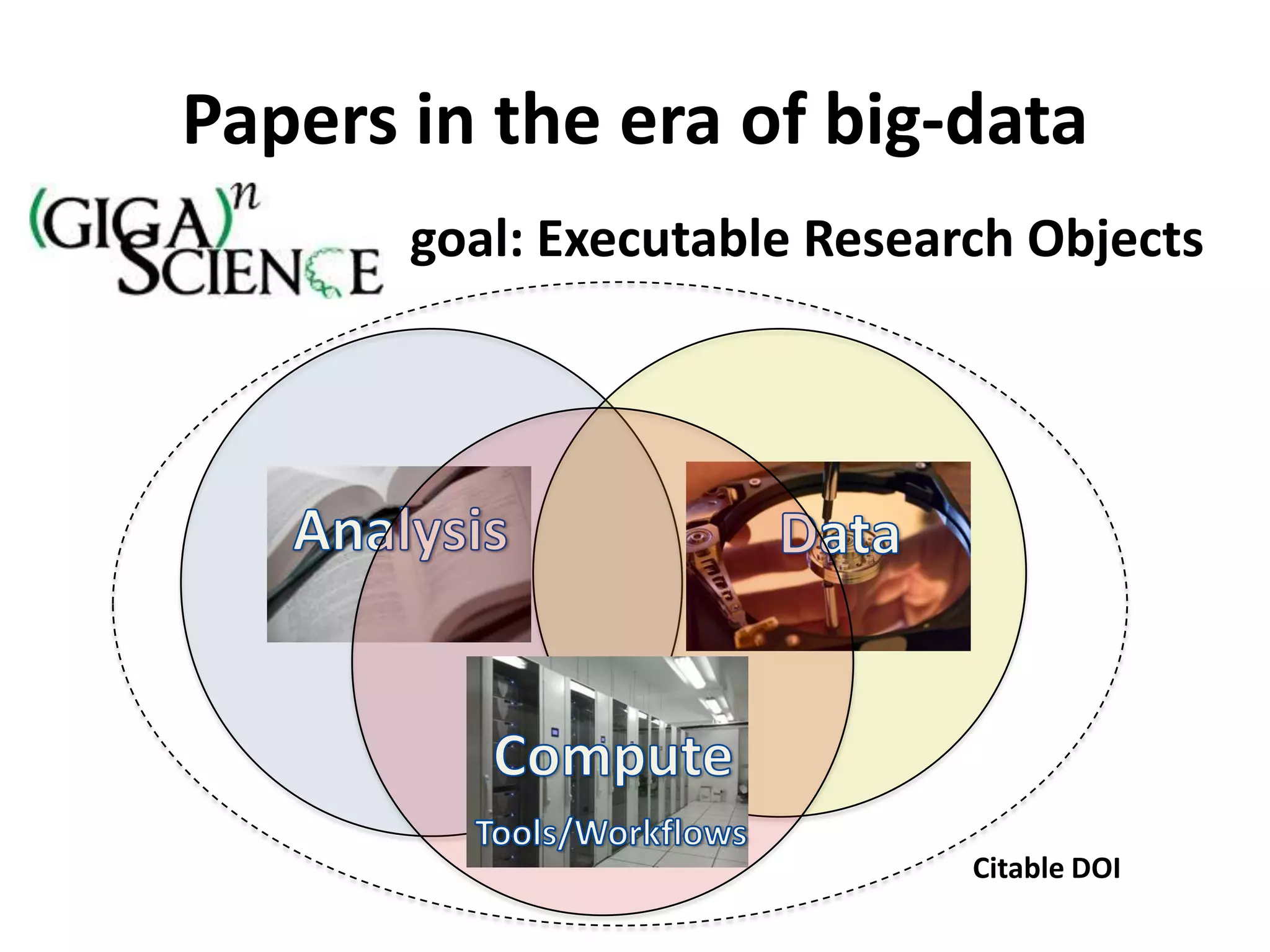
The Bio-IT World Asia meeting held on June 7, 2012, discussed challenges and advancements in managing and utilizing large-scale data, particularly in genomics, emphasizing the need for better data sharing practices and interoperability. It highlighted the importance of data citation through digital object identifiers (DOIs) to establish data as legitimate scholarly contributions and improve attribution for researchers. The discussions also touched upon the role of cloud solutions and the cultural hurdles in data sharing, advocating for incentives to share and publish data openly to facilitate collaborative research and public health responses.