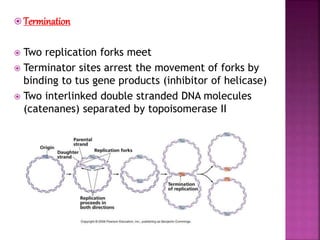
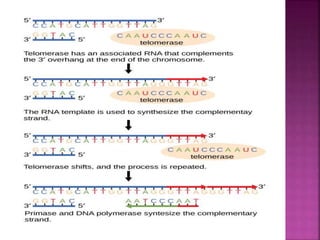
The document discusses the concept and process of DNA replication. It begins by defining replication as the copying of genetic information that is inherited to subsequent generations. It then summarizes the three proposed models of replication before discussing Matthew Meselson and Franklin Stahl's experiments, which supported the semiconservative model. The rest of the document details the specific mechanisms and steps of prokaryotic and eukaryotic replication, including initiation, elongation, termination, origins of replication, and the roles of various proteins. It concludes by discussing the importance of telomeres and telomerase in preventing chromosomal shortening during replication.