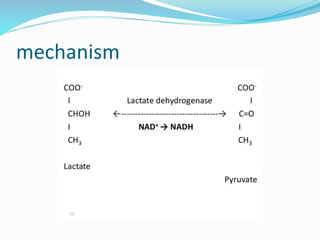
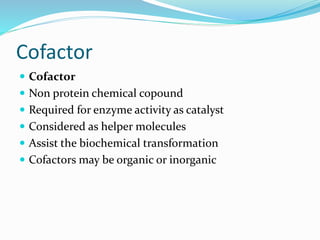
Cofactors are non-protein molecules that are required for enzyme activity and assist in biochemical transformations. They can be inorganic, such as metal ions like zinc, iron, and copper, or organic like vitamins. Organic cofactors, also called coenzymes, are low molecular weight organic compounds that participate in enzymatic reactions and include prosthetic groups that are tightly bound to proteins or cosubstrates that bind transiently. Common examples of coenzymes include nicotinamide adenine dinucleotide (NAD+) and vitamins. Cofactors activate apoenzymes and are essential for biological activity of enzymes.




![Iron –sulfur clusters
Iron-sulfur clusters are complexes of iron and sulfur atoms
held within proteins by cysteinyl residues.
They play both structural and functional roles, including
electron transfer, redox sensing, and as structural modules.
These clusters have many unique properties that are not
found in amino acids or other organic compounds.
[Fe-S] clusters participate in electron transfer, substrate
binding/activation, iron/sulfur storage, regulation of gene
expression, and enzyme activity
Definition
An iron-sulfur cluster is a unit comprising two or
more iron atoms and bridging sulfur ligands.](https://image.slidesharecdn.com/cofactorbindingqsdsd-191120184309/85/Cofactor-binding-bioinformatics-9-320.jpg)