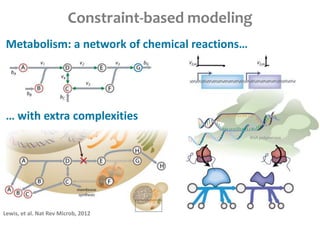
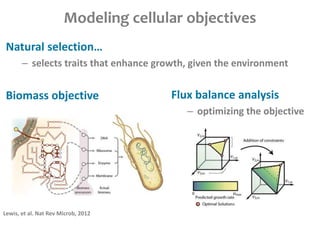
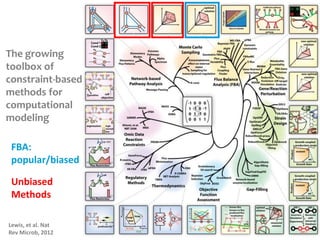
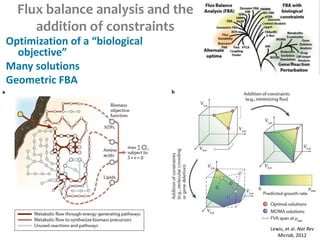
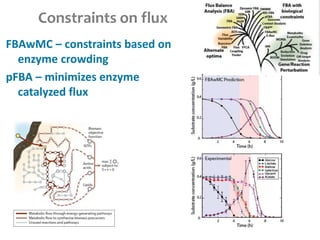
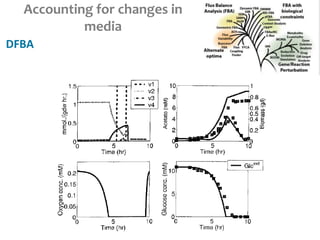
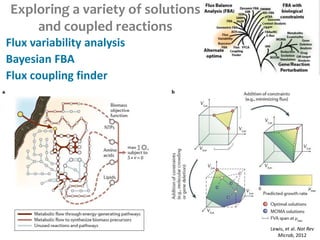
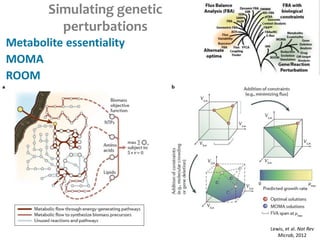
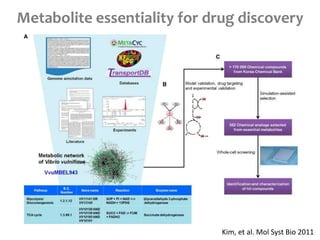
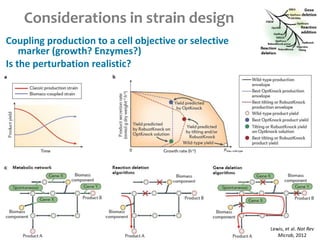
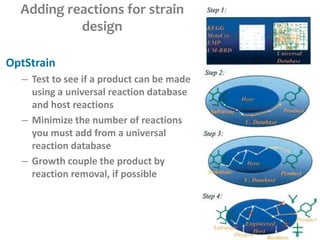
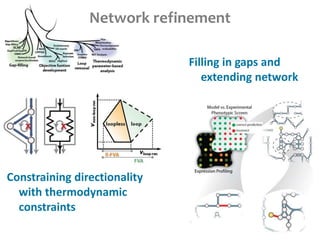
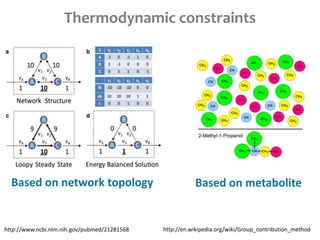
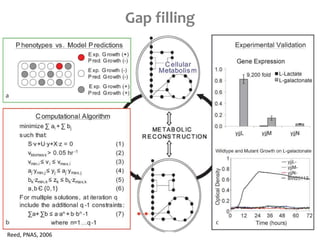
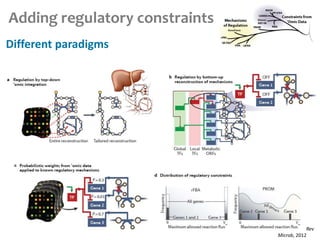
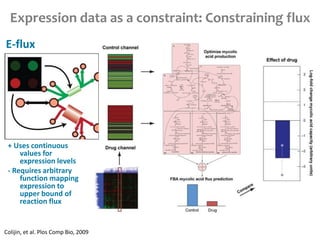
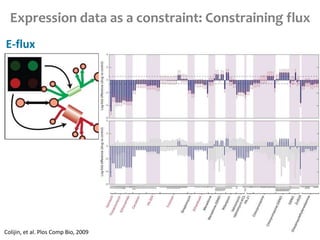
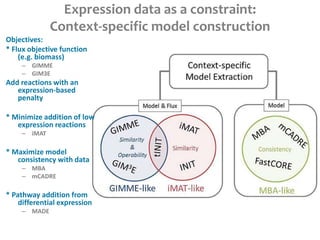
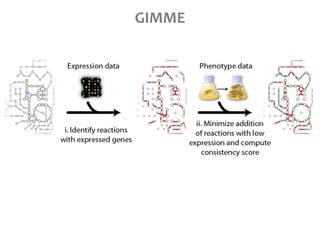
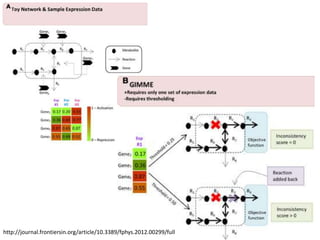
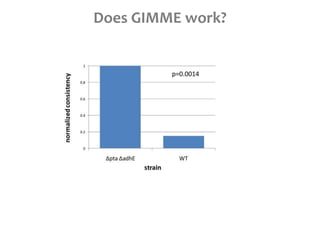
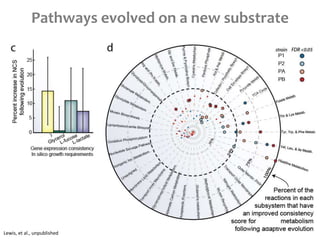
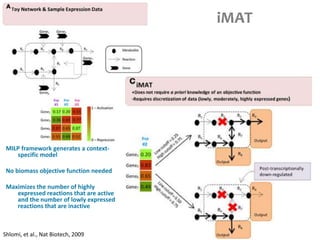
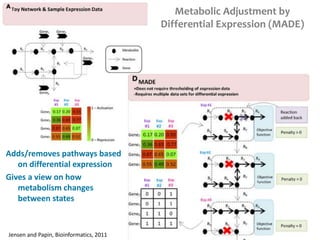
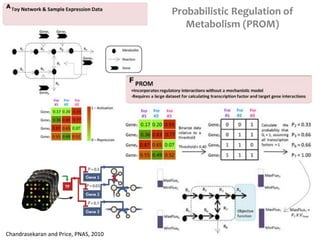
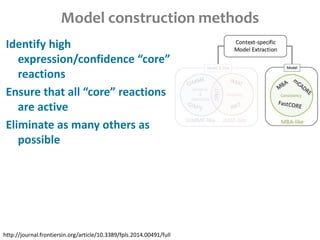
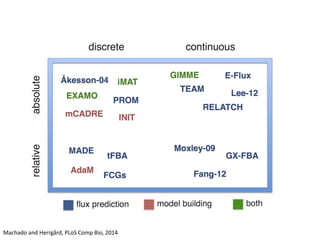
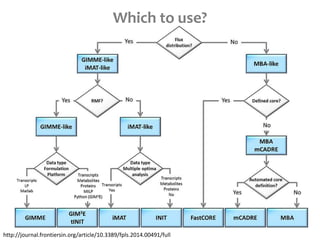
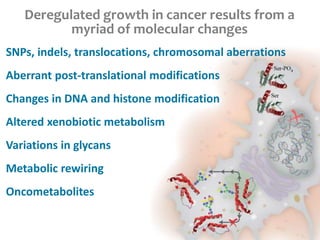
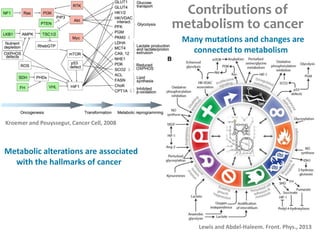
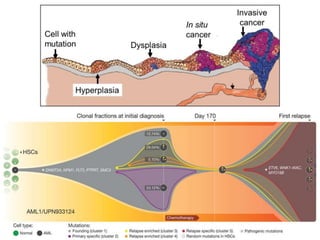
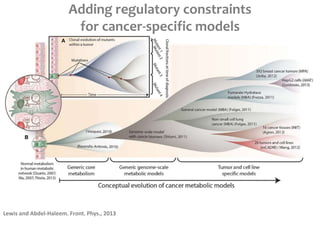
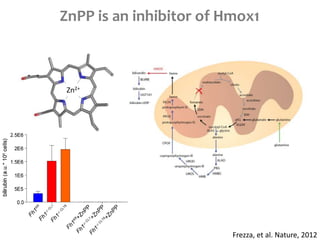
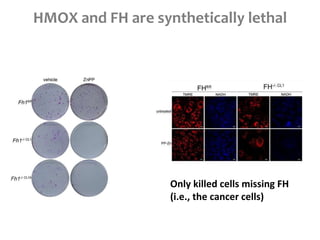
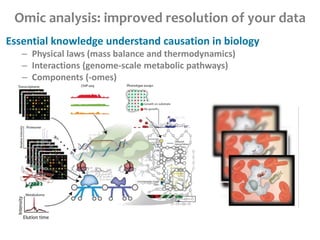
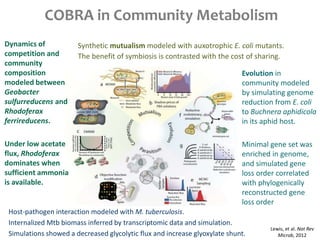
This document summarizes various constraint-based modeling methods for metabolic networks. It describes flux balance analysis and its extensions, which optimize an objective function like biomass production while incorporating additional constraints. Methods are presented that integrate gene expression data, thermodynamic constraints, and regulatory information to construct condition-specific metabolic models. Applications to cancer metabolism are discussed, such as identifying synthetic lethal drug targets. Community modeling examples are also provided that simulate microbial community dynamics and host-pathogen interactions.