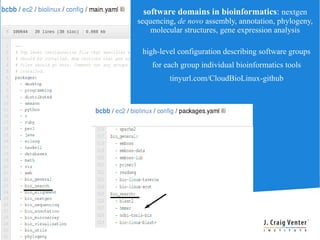
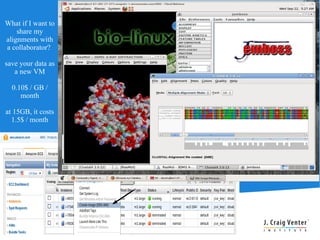
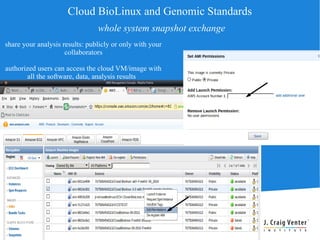
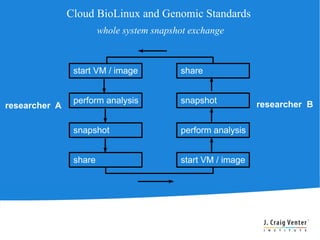
Cloud BioLinux is a standardized, pre-configured cloud computing solution for bioinformatics, facilitating on-demand access to genomic tools and resources. It allows users to create and share customized virtual machines (VMs) with a range of bioinformatics software, democratizing access to large-scale computing irrespective of institutional limitations. The initiative fosters collaboration within the community, aiming to expand user bases and improve software offerings while offering a rich, user-friendly interface for executing bioinformatics tasks.