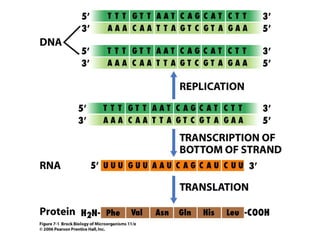
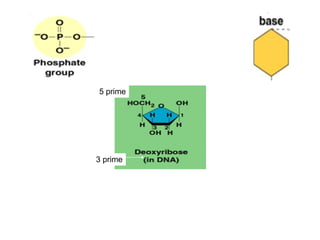
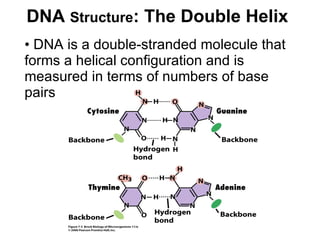
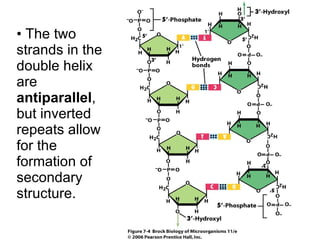
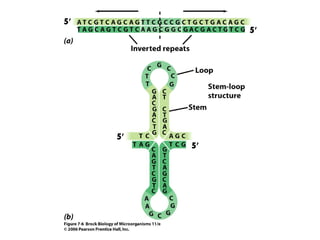
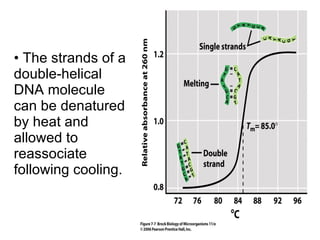
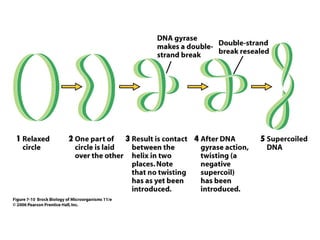
The document discusses key processes involved in gene expression and protein synthesis, including DNA replication, transcription, and translation. It provides details on:
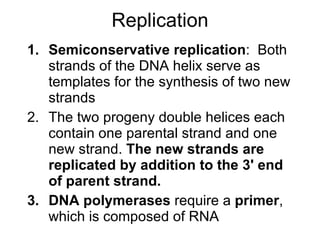
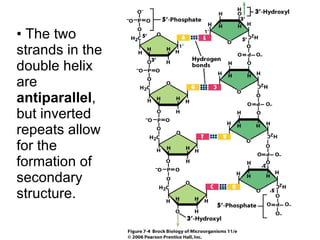
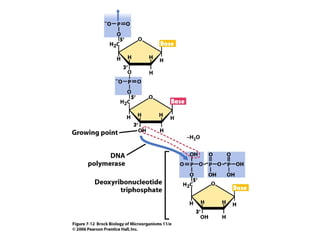
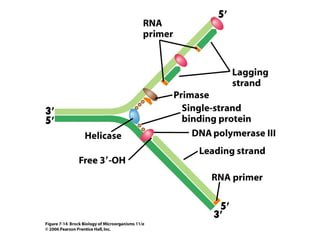
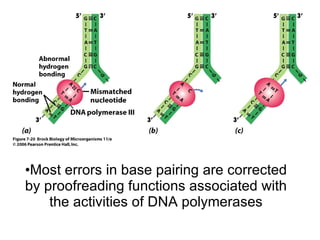
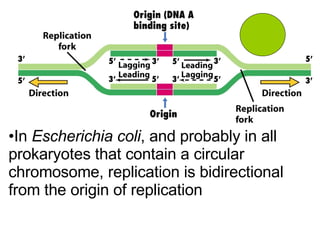
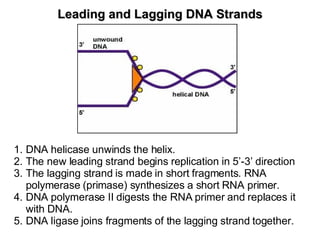
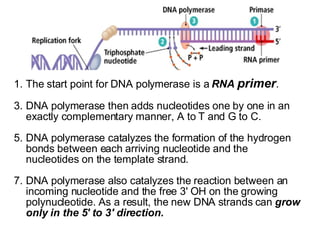
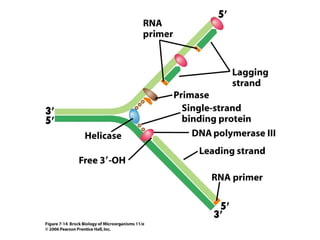
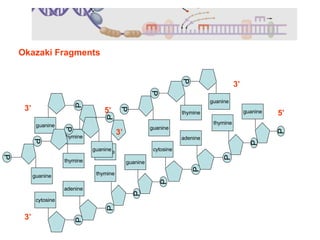
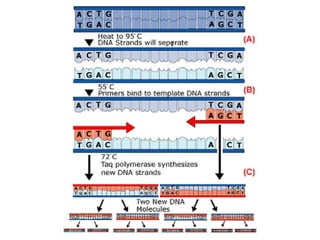
1) DNA replication through semiconservative replication where each new DNA double helix contains one original and one new strand synthesized in the 5' to 3' direction.
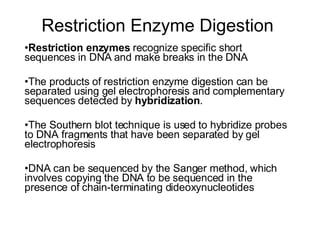
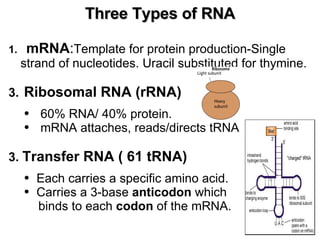
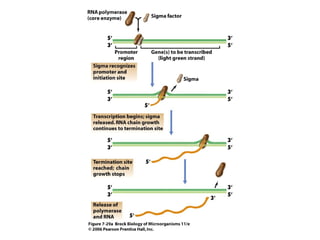
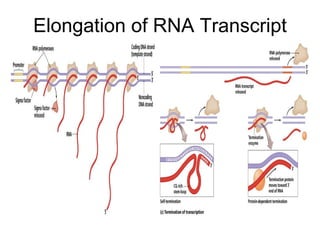
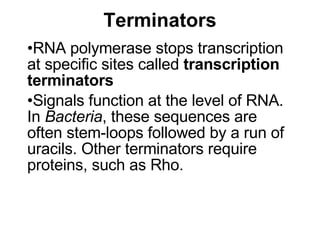
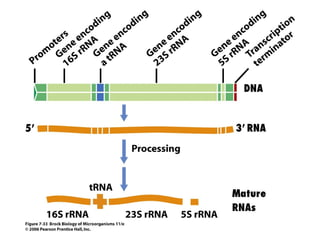
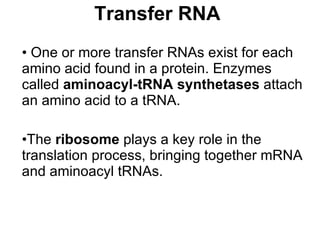
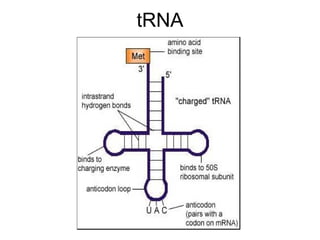
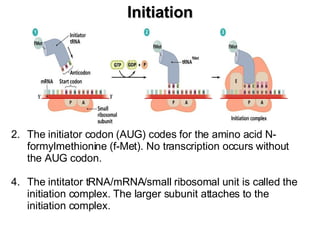
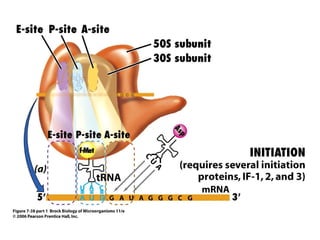
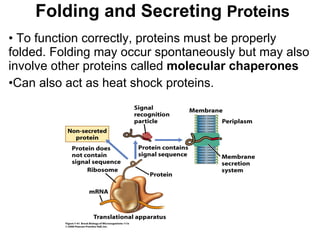
2) Transcription of DNA into mRNA which is then translated into proteins with the help of tRNA and the ribosome.
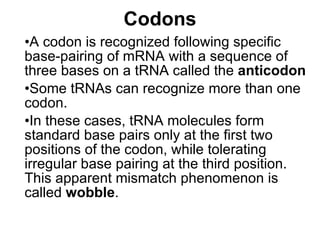
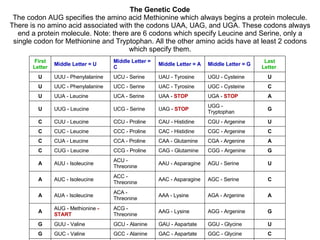
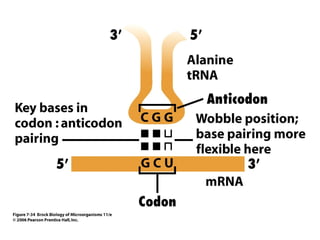
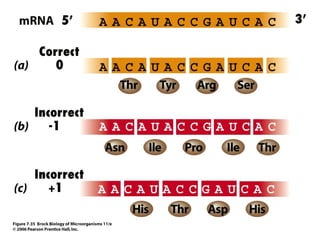
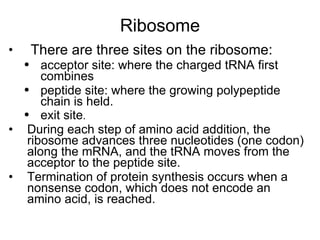
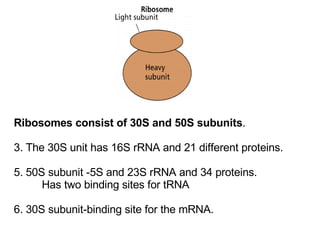
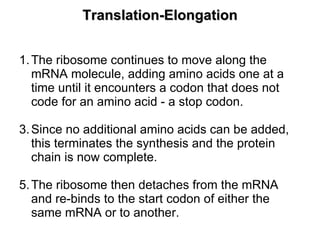
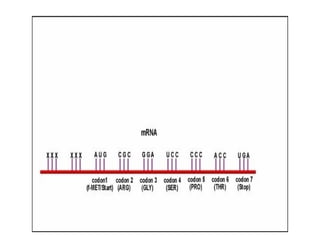
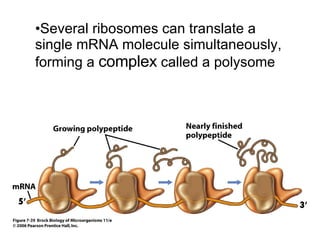
3) Translation of mRNA into polypeptide chains using the genetic code where codons in mRNA are recognized by anticodons in tRNA to add amino acids in the correct sequence. Translation terminates when a stop codon is reached.