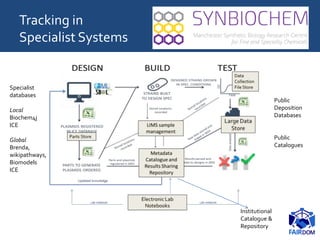
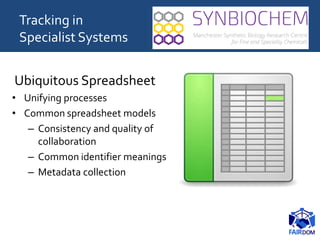
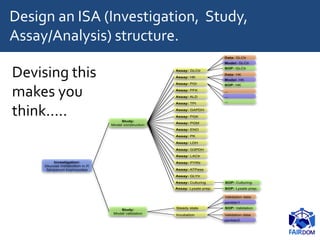
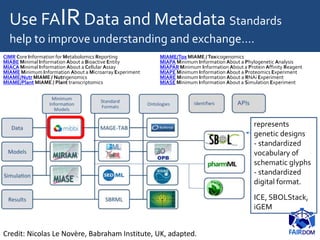
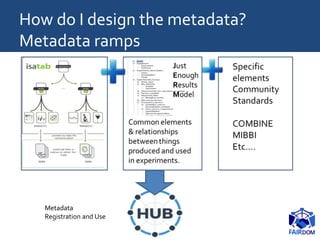
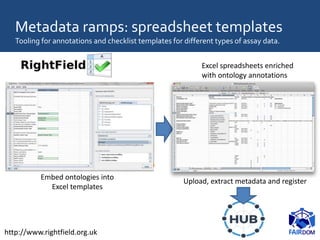
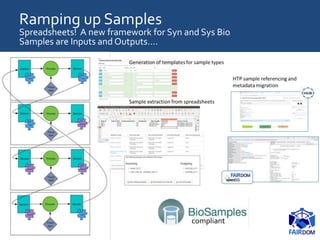
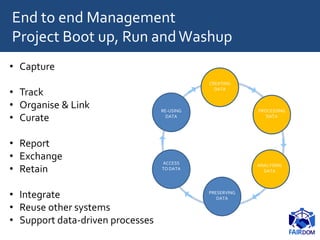
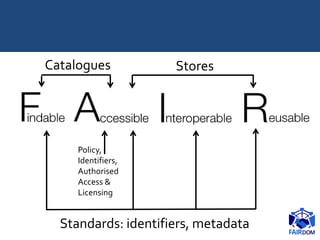
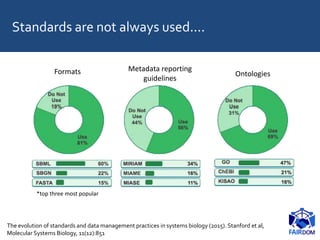
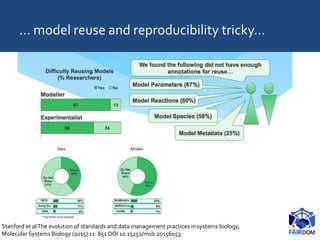
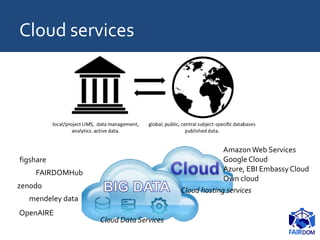
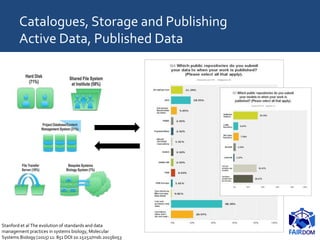
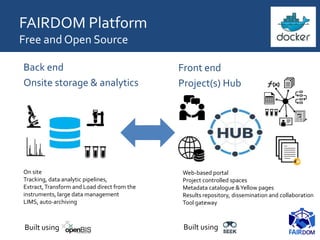
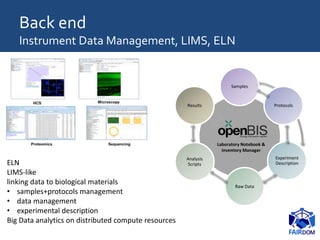
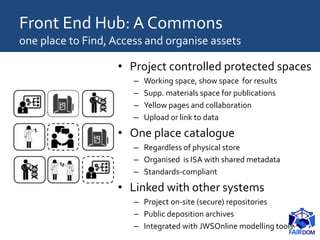
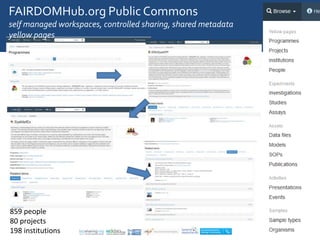
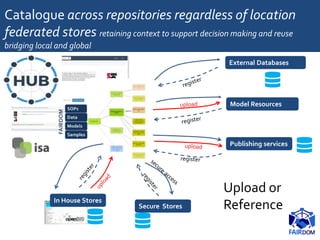
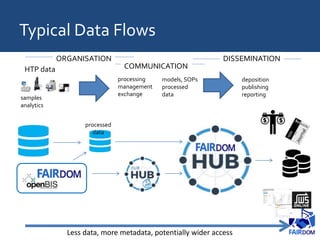
The document discusses the importance of fair data and model management in scientific research, particularly in systems and synthetic biology, emphasizing the need for effective data handling, documentation, and reproducibility. It outlines the FAIR principles (Findable, Accessible, Interoperable, Reusable) and presents strategies for automating data management processes, ensuring proper documentation, and maintaining collaboration across disciplines. Additionally, it highlights community efforts and resources aimed at improving data stewardship and the sharing of research outputs.



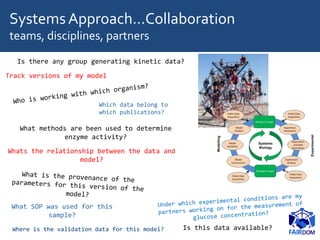
![SyntheticApproach - Automation
• Automate data management
– Spreadsheets, instruments, LIMS…
– Replication, comparison
• Support automation
– Tracking successful products from plasmids
– Informing robots
– Incorporate into pipelines and workflows
– Mediate through samples
– Standards
[Courtesy: Andrew Millar]](https://image.slidesharecdn.com/sssbs-lecture1-beingfair-goble-170718111056/85/Being-FAIR-FAIR-data-and-model-management-SSBSS-2017-Summer-School-6-320.jpg)















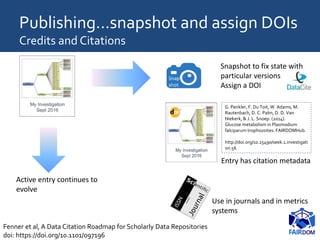
![18/07/2017 44
An “evolving manuscript” would begin with a pre-
publication, pre-peer review “beta 0.9” version of an
article, followed by the approved published article itself, [
… ] “version 1.0”.
Subsequently, scientists would update this paper with
details of further work as the area of research develops.
Versions 2.0 and 3.0 might allow for the “accretion of
confirmation [and] reputation”.
Ottoline Leyser […] assessment criteria in science revolve
around the individual. “People have stopped thinking
about the scientific enterprise”.
http://www.timeshighereducation.co.uk/news/evolving-manuscripts-the-future-of-scientific-communication/2020200.article](https://image.slidesharecdn.com/sssbs-lecture1-beingfair-goble-170718111056/85/Being-FAIR-FAIR-data-and-model-management-SSBSS-2017-Summer-School-44-320.jpg)
![Retention: Moses
from the ERANET SysMO Programme
Project ended in 2010
Publication in 2014/2015
Using data from 2012
[Maxim Zakhartsev]](https://image.slidesharecdn.com/sssbs-lecture1-beingfair-goble-170718111056/85/Being-FAIR-FAIR-data-and-model-management-SSBSS-2017-Summer-School-45-320.jpg)
![[Adapted from Ursula Klingmüller, Martin Böhm]
Excemplify
Antibody
Database
FAIR collaboration
from the ERANet ERASysAPP](https://image.slidesharecdn.com/sssbs-lecture1-beingfair-goble-170718111056/85/Being-FAIR-FAIR-data-and-model-management-SSBSS-2017-Summer-School-46-320.jpg)
![47
Programme
Overarching research theme (The Digital Salmon)
Project
Research grant (DigiSal, GenoSysFat)
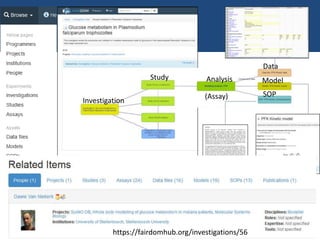
Investigation
A particular biological process, phenomenon or thing
(typically corresponds to [plans for] one or more closely related
papers)
Study
Experiment whose design reflects a specific biological research
question
Assay
Standardized measurement or diagnostic experiment using a
specific protocol
(applied to material from a study)
Jon Olav Vik,
Norwegian University of Life Science
Integration with Norway’s national
einfrastructure for Life Science (NeLS)](https://image.slidesharecdn.com/sssbs-lecture1-beingfair-goble-170718111056/85/Being-FAIR-FAIR-data-and-model-management-SSBSS-2017-Summer-School-47-320.jpg)