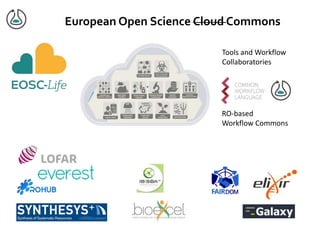
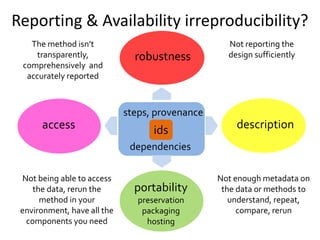
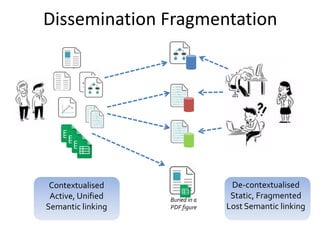
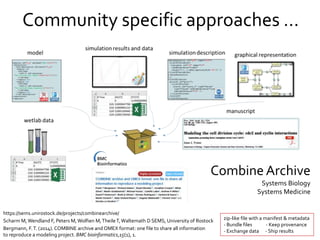
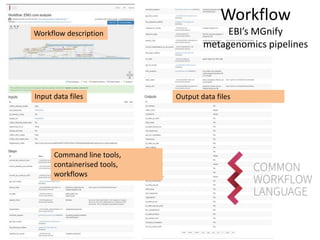
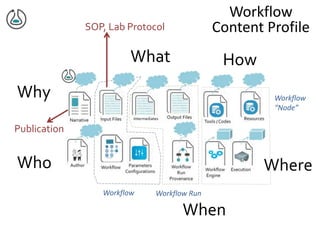
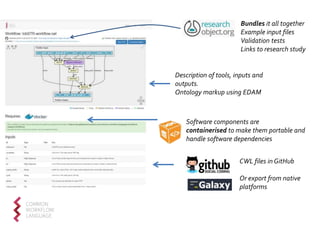
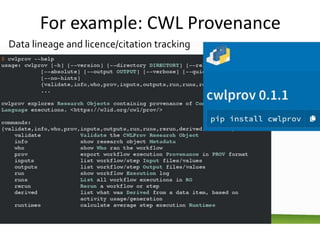
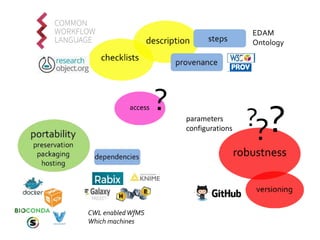
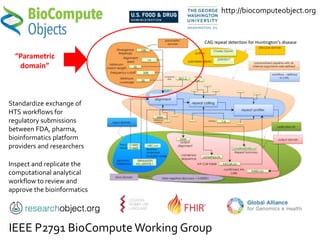
The document discusses the challenges of reproducible research in both wet and dry lab sciences, emphasizing the need for detailed reporting of methods, protocols, and data access. It presents solutions such as research objects that bundle together all necessary components—metadata, scripts, and software—facilitating transparency, portability, and interoperability in computational workflows. Finally, it highlights ongoing initiatives and tools, including workflow descriptions and containerization, to enhance the sharing and reproducibility of scientific research.



![Scientific publications
• announce a result
• convince readers to trust it
• enable a peer to reuse it or compare against it.
Wet Lab Experimental science
• describe the results
• provide a clear enough
description of the materials and
protocol to allow successful
repetition and extension [Jill
Mesirov 2010]
Dry Lab Computational science
• describe the results
• provide the complete software
development environment,
data, instructions, techniques
[David Donoho 1995]
Why?](https://image.slidesharecdn.com/gsc2019-goble-publish-190529151221/85/Reproducible-Research-how-could-Research-Objects-help-4-320.jpg)




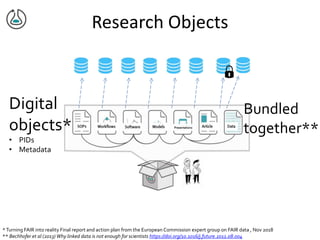
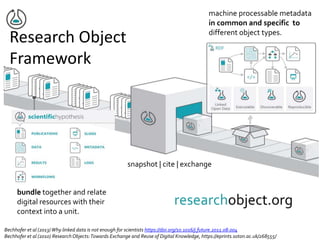
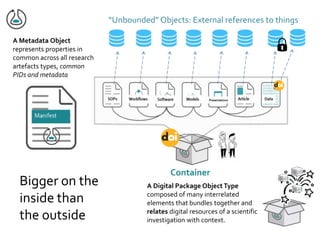
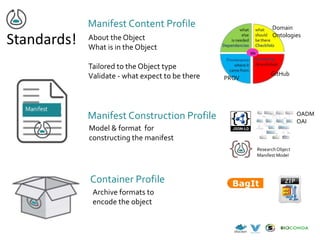



![NIH Data Commons
Big data distributed over multiple locations,
Efficiently and safely moved on demand
ROs are verified collections of references
[Chard, et al 2016]](https://image.slidesharecdn.com/gsc2019-goble-publish-190529151221/85/Reproducible-Research-how-could-Research-Objects-help-26-320.jpg)