Embed presentation
Downloaded 27 times














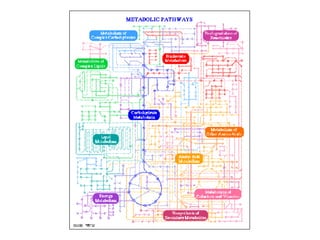


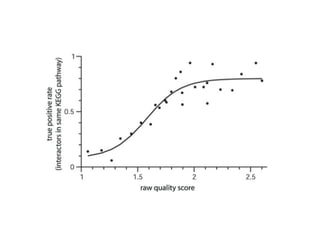












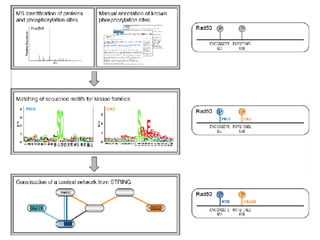


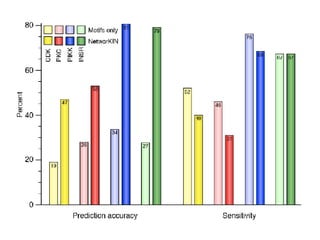

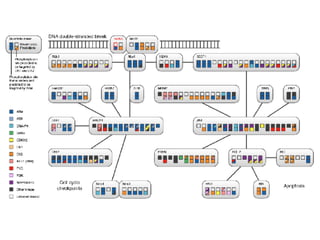











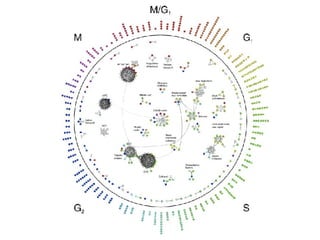


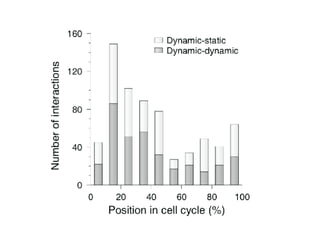

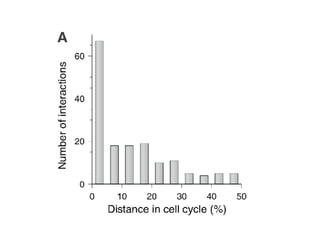




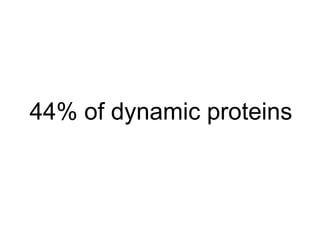


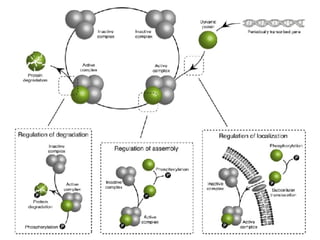







The document discusses the bioinformatics of cellular processes, focusing on protein networks, signaling, and regulation through methods like data integration and literature mining. It highlights the dynamics of protein interactions, the role of various proteins in cell cycle regulation, and benchmarking for computational network predictions. Additionally, it acknowledges contributions from various researchers and tools in advancing understanding in this field.