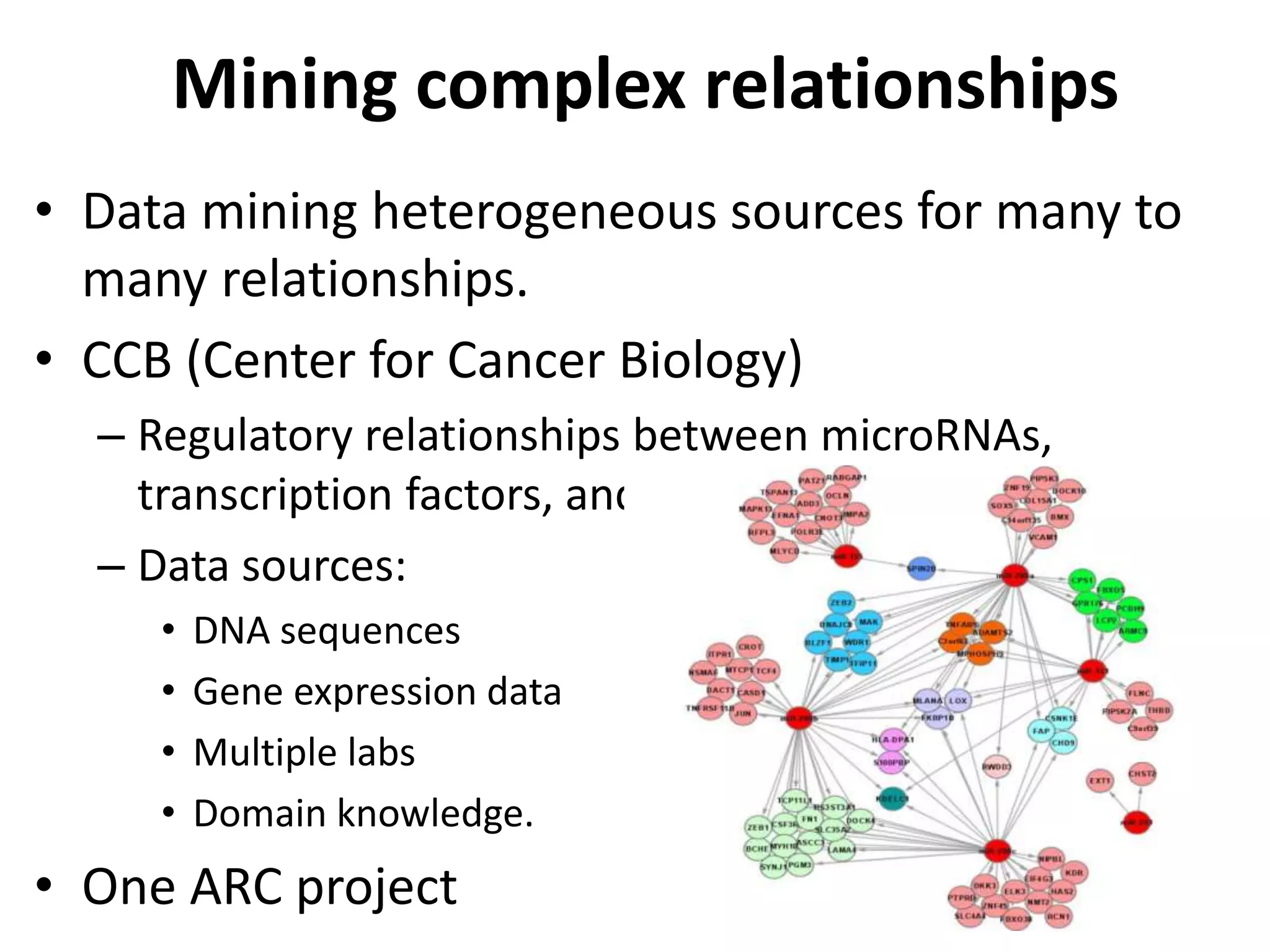
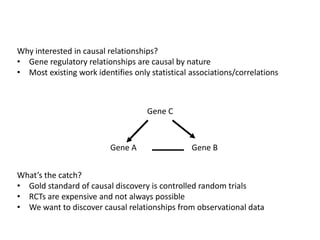
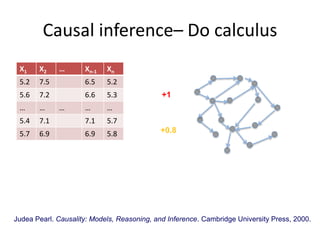
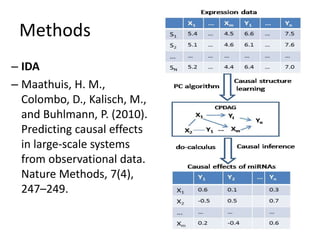
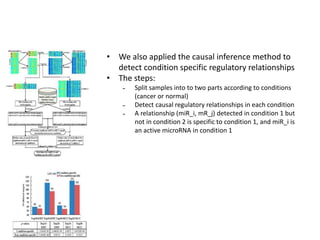
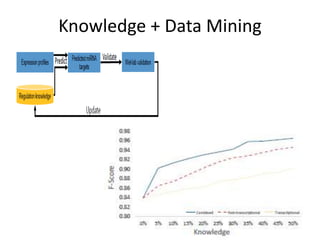
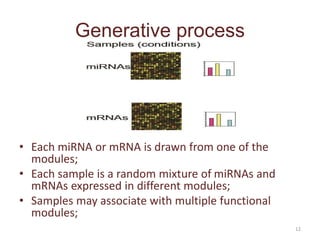
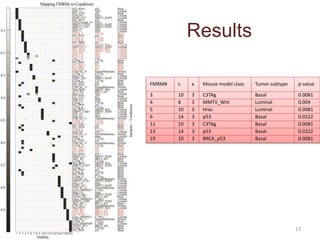
This document discusses mining complex relationships between microRNAs, transcription factors, and genes from heterogeneous data sources using causal inference approaches. Specifically, it describes a project that aims to infer regulatory relationships between microRNAs and mRNAs from multiple data sources including DNA sequences, gene expression data, and domain knowledge. It also discusses using causal inference methods like IDA to detect condition-specific regulatory relationships by analyzing samples split according to normal or cancer conditions.