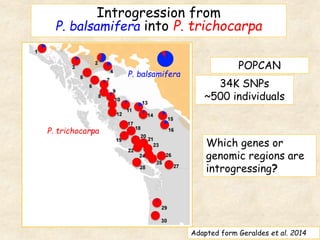
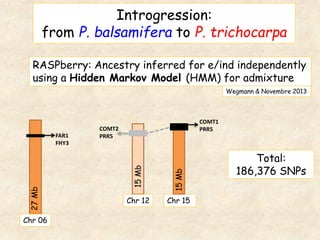
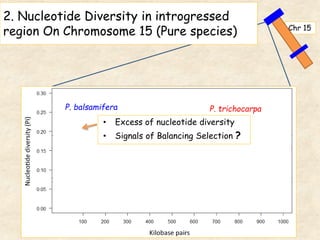
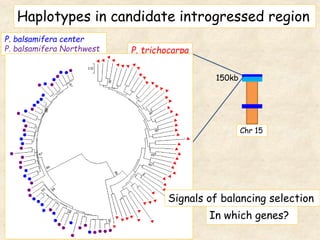
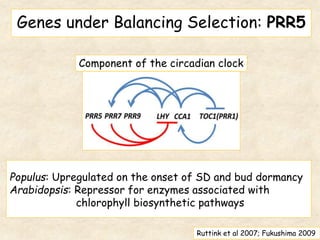
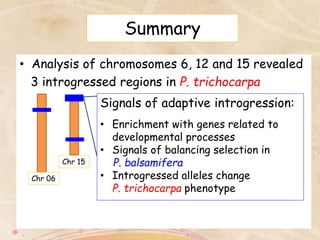
This study analyzed introgression from P. balsamifera into P. trichocarpa and found evidence that introgressed regions may provide adaptations. Three introgressed regions were identified on chromosomes 6, 12, and 15. The region on chromosome 15 showed enrichment for genes involved in development and signals of balancing selection, suggesting adaptive introgression. Introgressed alleles in this region were associated with higher chlorophyll content in P. trichocarpa, which could confer faster growth. Further genome-wide and phenotypic analyses are needed to better understand the adaptive effects of introgression.